| Concept | Frequentist | Bayesian |
|---|---|---|
| Parameter | Fixed | Random |
| Data | Random | Fixed |
| Estimate | Point estimate | Posterior distribution |
| Confidence Interval | Interval with coverage | Credible interval |
| Hypothesis Test | Reject/Fail to reject | Posterior predictive check |
| P-value | Probability of data under null | Probability of hypothesis given data |
| Likelihood | Function of parameter given data | Function of data given parameter |
Lecture 04: Population Structure and Bayesian Methods in Statistical Genetics
PUBH 8878, Statistical Genetics
Intro to Bayesian Methods
The Bayesian Paradigm
- Let \theta be unknown parameters, y observed data
- While frequentist methods treat \theta as fixed but unknown, Bayesian methods treat \theta as random variables

Sir Thomas Bayes
Bayes’ Theorem
- Uncertainty in \theta is modeled via a prior distribution p(\theta)
- Observed data y are modeled via a likelihood p(y|\theta)
- Bayes’ theorem combines these to yield the posterior distribution p(\theta|y) \propto p(y|\theta)p(\theta)
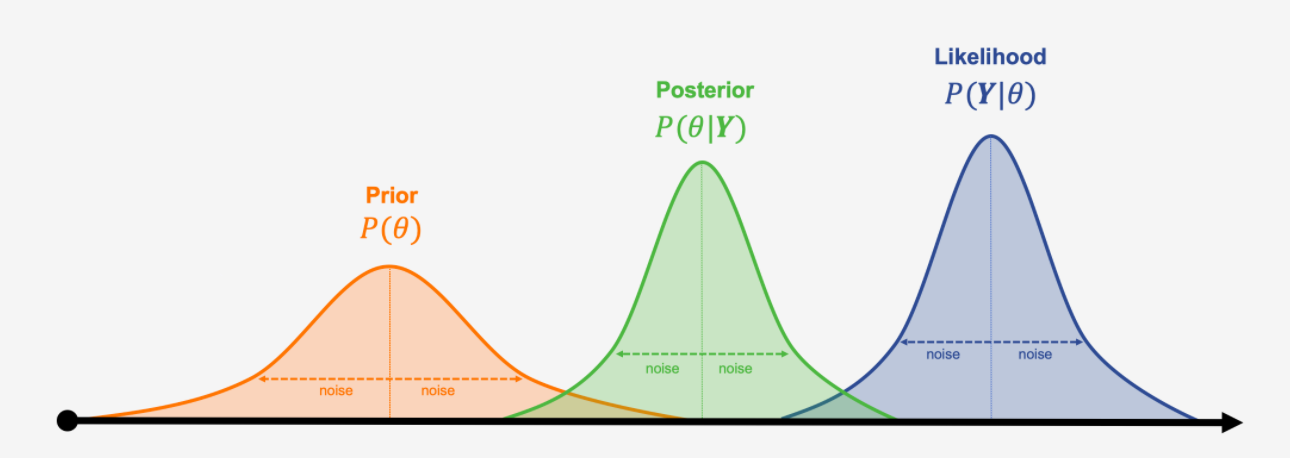
Frequentist vs Bayesian Terminology
Interpretation of Intervals
Compare statements on intervals
- Frequentist: “Over many repeated samples, 95% of such intervals will contain the true parameter.”
- Bayesian: “Given the observed data and prior, there is a 95% probability that the parameter lies within this interval.”
Bayesian Pros and Cons
Pros
- Intuitive: direct probability statements about parameters
- Flexibility: complex models, hierarchical structures, small-n
- Incorporate prior knowledge or expert opinion
- Full uncertainty quantification via posterior distributions
Cons
- Computationally intensive: MCMC, variational inference
- Sensitivity to prior choice
- Not the status quo in many fields
Further Resources
Conjugacy
- A prior is conjugate to a likelihood if the posterior is in the same family as the prior.
- Example: Beta prior + Binomial likelihood to Beta posterior
Conjugacy Example
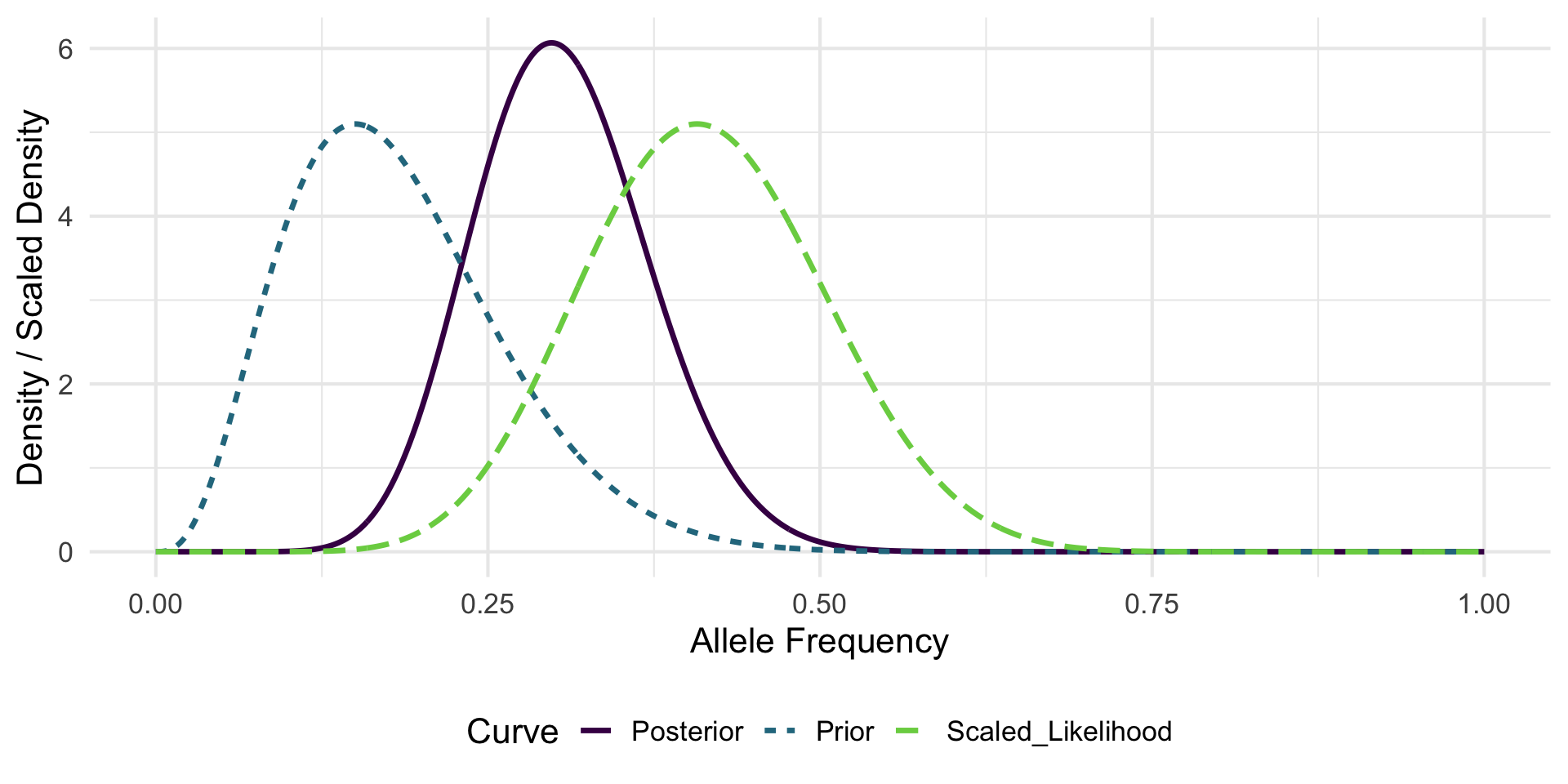
Consider the allele-frequency estimation setup introduced in Lecture 1.
- Previous experiment: observed x_{\text{prev}} = 3 successes (allele A) out of n_{\text{prev}} = 20 trials.
- Current data: x = 11 successes out of n = 27 trials.
- Parameter of interest: allele (“success”) frequency p.
Step 1: Prior
From the previous experiment (3 successes out of 20 trials), we can form a Beta prior:
Interpret \alpha and \beta as pseudo-counts of successes and failures, respectively. (Note that \text{Beta}(1,1) is uniform on 0,1.)
\begin{gather*} p \sim \text{Beta}(\alpha, \beta) \\ \alpha = x_{\text{prev}} + 1 = 4 \\ \beta = n_{\text{prev}} - x_{\text{prev}} + 1 = 18. \end{gather*}
Step 2: Likelihood
Current data likelihood (up to proportionality in p): L(p; x,n) \propto p^{x} (1-p)^{n-x} = p^{11}(1-p)^{16}.
Recognize this kernel as
\text{Beta}(x+1, n-x+1)=\text{Beta}(12,17)
Step 3: Posterior (Conjugacy)
Multiply prior and likelihood kernels: p^{\alpha-1}(1-p)^{\beta-1}\times p^{x}(1-p)^{n-x} = p^{(\alpha + x) - 1}(1-p)^{(\beta + n - x) - 1}.
Thus
\begin{align*} p | x \sim \text{Beta}(\alpha + x,\ \beta + n - x) &= \text{Beta}(4+11,\ 18+16) \\ &= \text{Beta}(15,34). \end{align*}
Posterior Summaries
Posterior mean: E[p|x] = \frac{15}{15+34} = 0.306 (shrinks slightly toward prior mean \frac{4}{22}=0.182 relative to MLE \hat p = 11/27 \approx 0.407).
Effective sample size intuition: prior contributes (\alpha+\beta-2)=20 pseudo-trials; data contribute 27 real trials.
Visualization
Non-Conjugate Models
- Conjugacy is convenient but limited to simple models
- Many realistic models (e.g., multinomial/Dirichlet with nonlinear transforms) are non-conjugate
- Bayesian inference in these models typically requires computational methods like MCMC or variational inference
- Richard McElreath has a great youtube video on MCMC
Bayesian Revisit: ABO Allele Frequencies
- Goal: Infer allele frequencies
(p_A, p_B, p_O)given phenotype counts(n_A, n_AB, n_B, n_O). - Frequentist (Lecture 03): EM treats latent genotypes for A and B phenotypes.
- Bayesian: Place a prior on allele frequencies; integrate (average) over uncertainty rather than impute expected counts.
Data & Sufficient Statistics
- From Lecture 03, we have:
- n_A = 725 individuals with A phenotype
- n_{AB} = 72 individuals with AB phenotype
- n_{B} = 258 individuals with B phenotype
- n_{O} = 1073 individuals with O phenotype
- Total sample size: N = 2128 individuals
Model Specification
- Allele frequency vector: \boldsymbol p=(p_A,p_B,p_O), \boldsymbol p\sim\text{Dirichlet}(\boldsymbol\alpha).
- Under HWE, genotype frequencies: p_A^2, 2p_A p_O, p_B^2, 2 p_B p_O, 2 p_A p_B, p_O^2.
- Phenotype probabilities (aggregating ambiguous genotypes):
- P(\text{A}) = p_A^2 + 2 p_A p_O
- P(\text{AB}) = 2 p_A p_B
- P(\text{B}) = p_B^2 + 2 p_B p_O
- P(\text{O}) = p_O^2
- Likelihood: (n_A,n_{AB},n_B,n_O) \sim \text{Multinomial}(N, \boldsymbol q) with \boldsymbol q above.
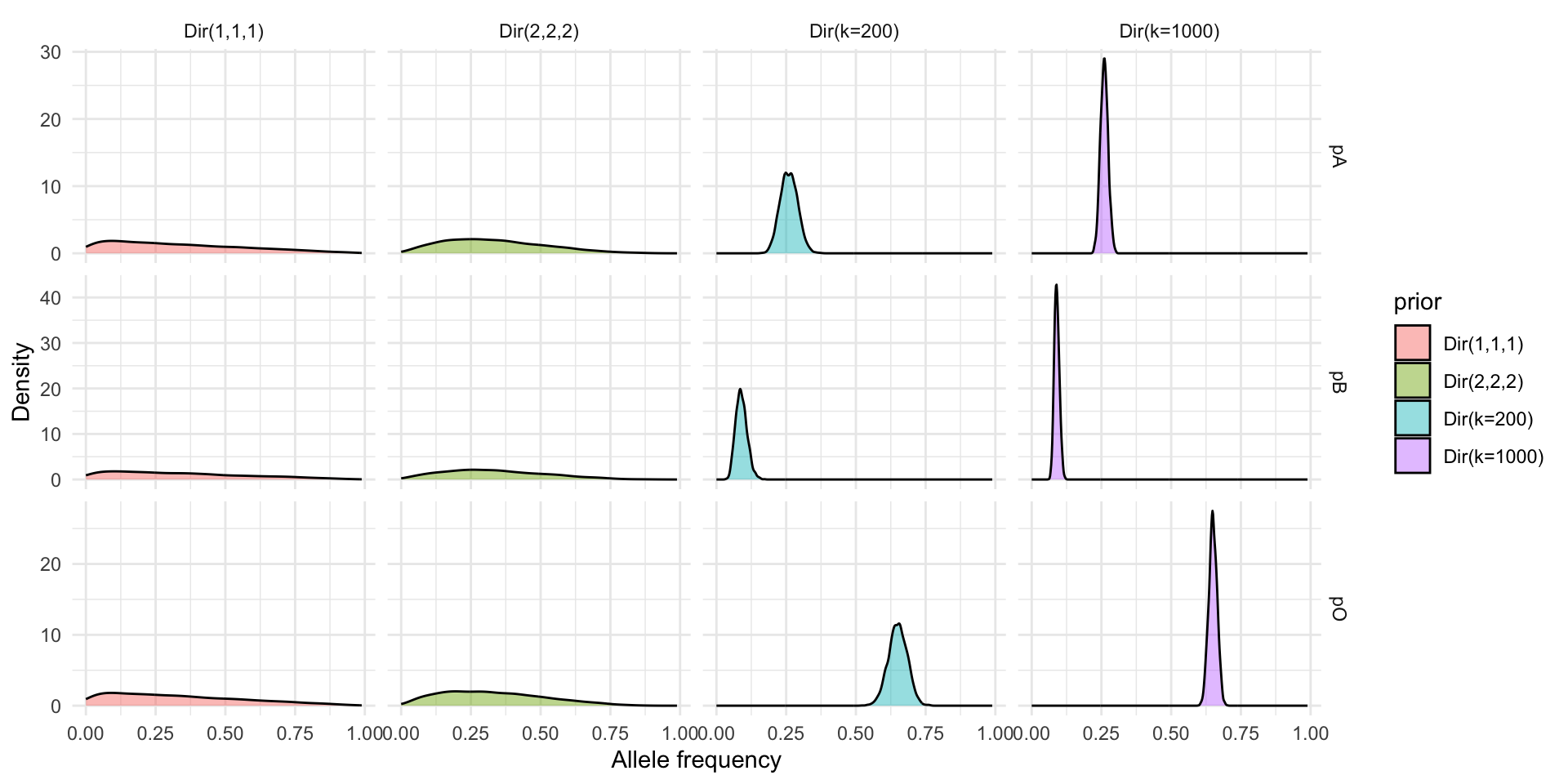
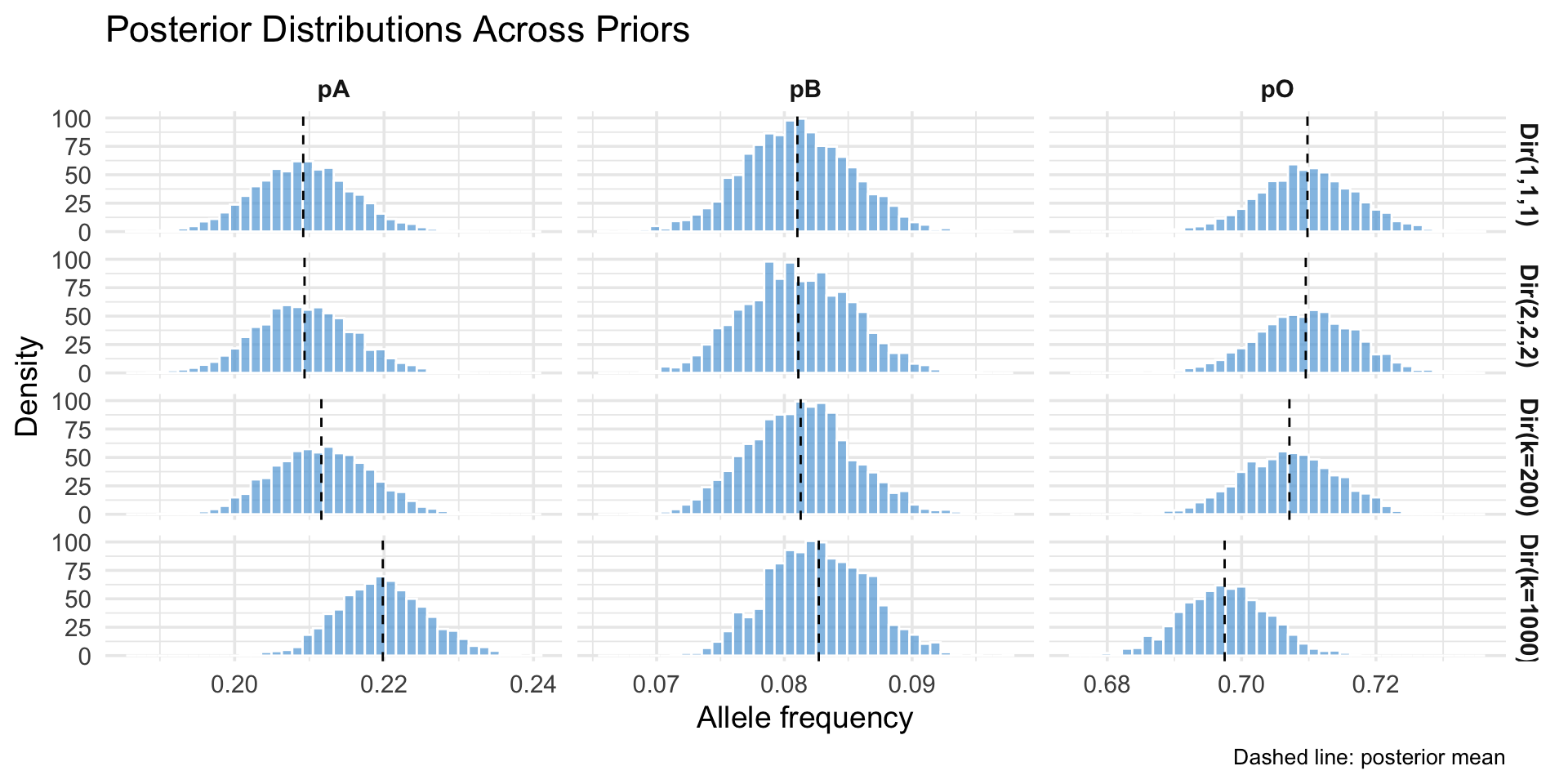
Prior Families
- Weak Dirichlet(1,1,1) (uniform over allele simplex)
- Mild Dirichlet(2,2,2) (light shrink toward center)
Using Historical Data
- Consider global survey means (p_A = 0.26, p_B = 0.09, p_O = 0.65) (Mourant et al., 1976; Yamamoto et al., 2012).
- Dirichlet prior: \mathbf{\alpha} = k (0.26, 0.09, 0.65)
- Effective sample size idea: acts like observing k allele draws before current data (2N alleles in sample).
- Relative weight vs data (here total alleles = 2N = 4256):
- k = 200 \to prior weight \approx 4.5\% of total information.
- k = 1000 \to prior weight \approx 19.0\% of total information.
Prior Shapes (Marginal Densities)
How to fit these models?
- So we have our data, likelihood, and priors
- We can use MCMC to sample from the posterior distribution of allele frequencies
- There exist many software packages to do this! We will use Stan (Carpenter et al., 2017) via the
cmdstanrR package

Stan Model
Step 1: Phenotype Probabilities
- 1
-
Declare the
functionsblock (optional in Stan, but lets us encapsulate logic) - 2
- Define a helper that maps allele frequencies to phenotype probabilities
- 3
- Extract components (for readability in later expressions)
Stan Model
Step 1: Phenotype Probabilities
- 4
-
Allocate a length-4 vector
q(A, AB, B, O) for phenotype probabilities - 5
- Hardy–Weinberg genotype algebra aggregated into phenotype probabilities
- 6
- Return the vector
Stan Model
Step 2: Data & Transforms
- 1
-
Raw observed counts and prior hyperparameters enter in the
datablock. - 2
- ABO phenotype counts (non-negative integers) per category.
- 3
-
Dirichlet prior parameters supplied from R (allow different priors via
alpha).
Stan Model
Step 2: Data & Transforms
- 4
-
transformed datapre-computes deterministic quantities once - 5
- Total sample size N used for reference or diagnostics
- 6
- Assemble counts into an array to pass to the multinomial
Stan Model
Step 3: Parameters & Derived q
- 1
-
Declare unknown quantities to infer in
parameters - 2
-
simplex[3]enforces positivity and sum-to-one constraints automatically - 3
-
transformed parametersrecomputes per-draw derived values - 4
- Reuse helper to obtain phenotype probabilities from allele frequencies
Stan Model (Step 4: Prior & Likelihood)
- 1
-
The
modelblock contains all sampling statements contributing to log density - 2
-
Dirichlet prior on allele frequencies
p - 3
-
Multinomial likelihood over phenotype counts with probabilities
q.
Stan Model (Step 5: Generated Quantities)
1generated quantities {
2 real log_lik = multinomial_lpmf(y | q);
3 vector[6] geno_freq;
4 real pA = p[1]; real pB = p[2]; real pO = p[3];
5 geno_freq[1] = pA * pA; # AA
geno_freq[2] = 2 * pA * pO; # AO
geno_freq[3] = pB * pB; # BB
geno_freq[4] = 2 * pB * pO; # BO
geno_freq[5] = 2 * pA * pB; # AB
geno_freq[6] = pO * pO; # OO
}- 1
- Post-processing: quantities saved per posterior draw
- 2
- Store log-likelihood for model comparison / LOO / WAIC
- 3
- Allocate genotype frequency vector
- 4
- Local aliases improve clarity when computing genotype frequencies
- 5
- Hardy–Weinberg genotype probabilities
Compile Stan Model
- Stan uses C++ on the backend, so we need to compile the model once before fitting
cmdstan_model()handles compilation and returns a model object for sampling
Fit: Weak Prior (Dirichlet(1,1,1))
- 1
-
Execute the MCMC sampler using the
sample()method on the compiled model object - 2
- Pass data from R to Stan as a named list
- 3
- Specify MCMC sampler settings
Running MCMC with 4 parallel chains...
Chain 1 finished in 0.0 seconds.
Chain 2 finished in 0.0 seconds.
Chain 3 finished in 0.0 seconds.
Chain 4 finished in 0.0 seconds.
All 4 chains finished successfully.
Mean chain execution time: 0.0 seconds.
Total execution time: 0.2 seconds.Model Diagnostics
# A tibble: 18 × 10
variable mean median sd mad q5 q95 rhat ess_bulk
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 lp__ -2.31e+3 -2.31e+3 9.83e-1 7.26e-1 -2.31e+3 -2.31e+3 1.00 1880.
2 p[1] 2.09e-1 2.09e-1 6.60e-3 6.52e-3 1.99e-1 2.20e-1 1.00 4495.
3 p[2] 8.10e-2 8.09e-2 4.23e-3 4.26e-3 7.41e-2 8.81e-2 1.00 2125.
4 p[3] 7.10e-1 7.10e-1 7.27e-3 7.18e-3 6.98e-1 7.22e-1 1.00 3368.
5 q[1] 3.41e-1 3.41e-1 9.80e-3 9.84e-3 3.25e-1 3.57e-1 1.00 4376.
6 q[2] 3.39e-2 3.39e-2 1.92e-3 1.92e-3 3.08e-2 3.70e-2 1.00 2338.
7 q[3] 1.22e-1 1.21e-1 6.27e-3 6.21e-3 1.11e-1 1.32e-1 1.00 2210.
8 q[4] 5.04e-1 5.04e-1 1.03e-2 1.02e-2 4.87e-1 5.21e-1 1.00 3368.
9 log_lik -1.16e+1 -1.13e+1 9.81e-1 7.14e-1 -1.36e+1 -1.07e+1 1.00 1867.
10 geno_freq… 4.38e-2 4.37e-2 2.76e-3 2.73e-3 3.94e-2 4.85e-2 1.00 4495.
11 geno_freq… 2.97e-1 2.97e-1 7.10e-3 7.08e-3 2.85e-1 3.08e-1 1.00 4242.
12 geno_freq… 6.58e-3 6.55e-3 6.87e-4 6.90e-4 5.50e-3 7.76e-3 1.00 2125.
13 geno_freq… 1.15e-1 1.15e-1 5.59e-3 5.53e-3 1.06e-1 1.24e-1 1.00 2225.
14 geno_freq… 3.39e-2 3.39e-2 1.92e-3 1.92e-3 3.08e-2 3.70e-2 1.00 2338.
15 geno_freq… 5.04e-1 5.04e-1 1.03e-2 1.02e-2 4.87e-1 5.21e-1 1.00 3368.
16 pA 2.09e-1 2.09e-1 6.60e-3 6.52e-3 1.99e-1 2.20e-1 1.00 4495.
17 pB 8.10e-2 8.09e-2 4.23e-3 4.26e-3 7.41e-2 8.81e-2 1.00 2125.
18 pO 7.10e-1 7.10e-1 7.27e-3 7.18e-3 6.98e-1 7.22e-1 1.00 3368.
# ℹ 1 more variable: ess_tail <dbl>Posterior Summaries & Intervals (Weak Prior)
post_weak <- fit_weak$draws(variables = c("p")) |> as_draws_df()
weak_summ <- post_weak %>% summarise(
mean_pA = mean(`p[1]`), mean_pB = mean(`p[2]`), mean_pO = mean(`p[3]`),
sd_pA = sd(`p[1]`), sd_pB = sd(`p[2]`), sd_pO = sd(`p[3]`)
)
weak_ci <- post_weak %>% summarise(
pA_low = quantile(`p[1]`, 0.025), pA_high = quantile(`p[1]`, 0.975),
pB_low = quantile(`p[2]`, 0.025), pB_high = quantile(`p[2]`, 0.975),
pO_low = quantile(`p[3]`, 0.025), pO_high = quantile(`p[3]`, 0.975)
)Posterior Histograms (Weak Prior)
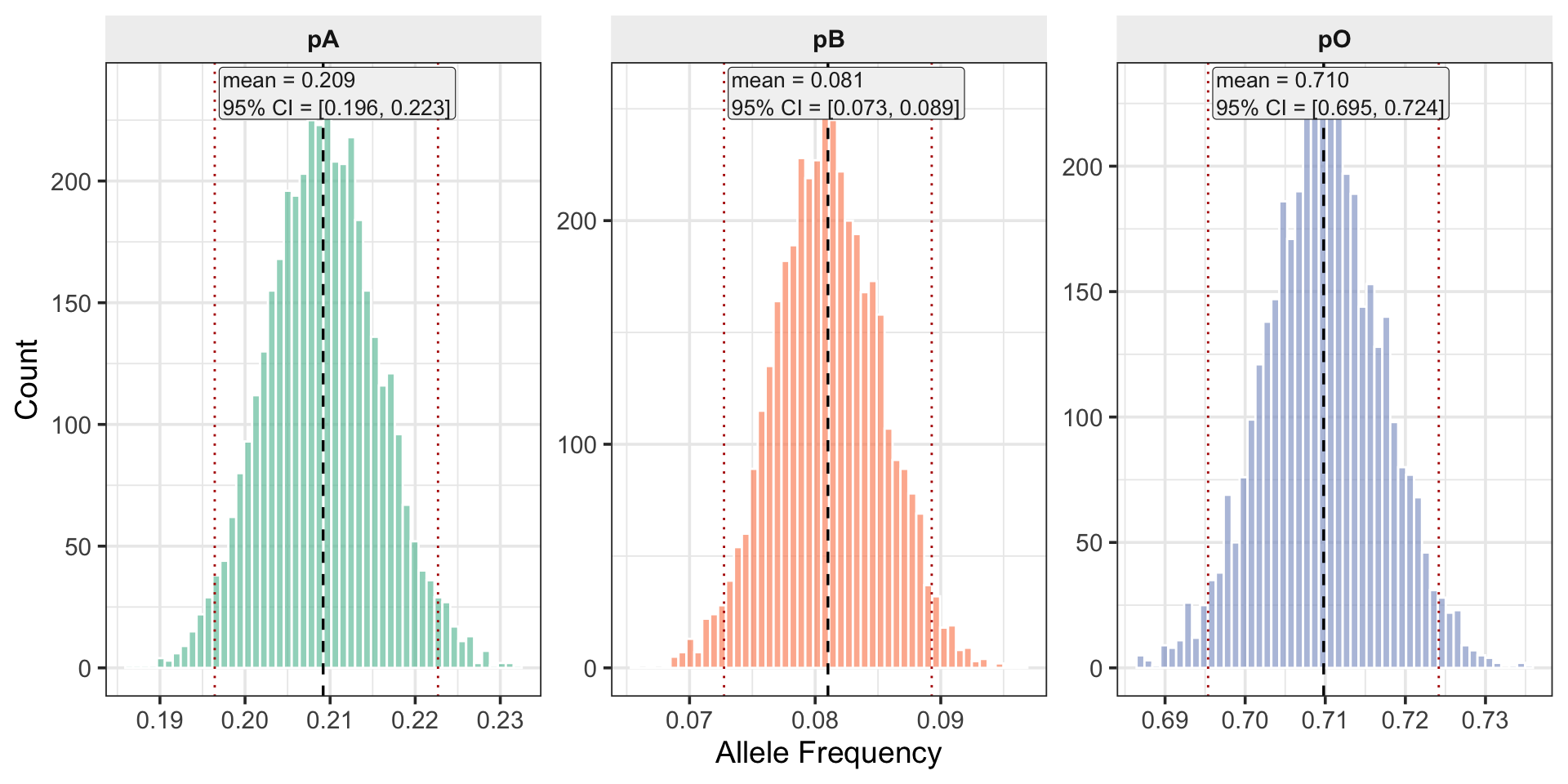
EM vs Bayesian Point Estimates
# A tibble: 2 × 5
method pA pB pO loglik
<chr> <dbl> <dbl> <dbl> <dbl>
1 EM 0.209 0.0808 0.710 -2304.
2 Posterior Mean 0.209 0.0810 0.710 NA Consolidated Posterior Comparison
Population Structure
Population Substructure
- Features of a population which result from variation of expected allele frequencies across individuals
- Standard allele counting (\hat{p} = (2n_{AA} + n_{Aa}/2n)) will still be unbiased
- But, not all subjects may have the same probability of being represented in the sample
- Variance of estimate will be effected
Population Stratification
- Individuals in a population can be subdivided into mutually exclusive strata
- Within each strata the allele frequency is the same for all individuals
- Intuitively, we are partitioning a large dataset into multiple smaller datasets
Population Admixture
- When individuals in a population have a mixture of different genetic ancestries due to prior mixing of two or more populations
- Often result of migration
Population Admixture
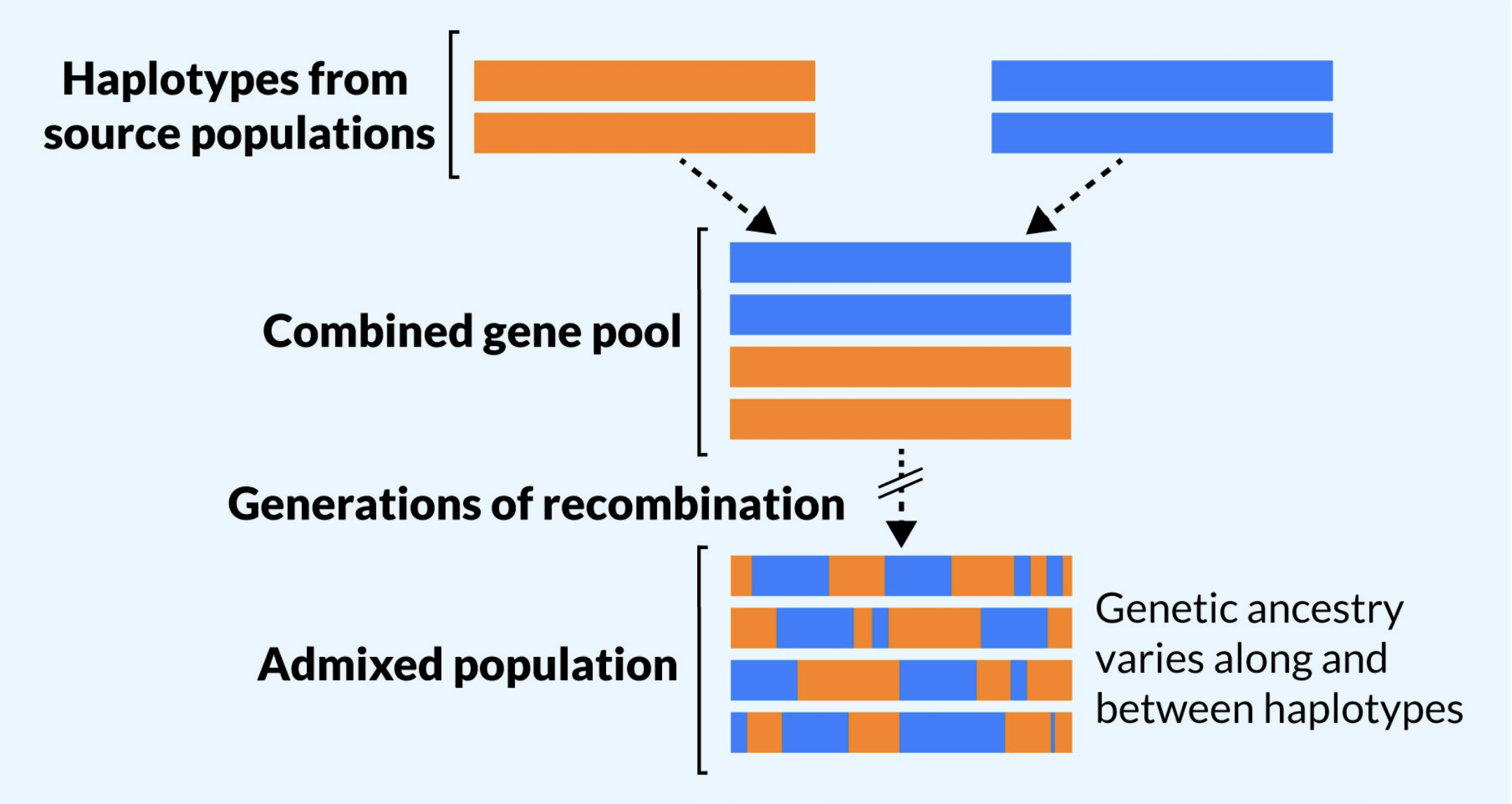
From Korunes and Goldberg (2021)
Population Inbreeding
- Occurs when there is a preference for mating among relatives in a population or because geographic isolation of subgroups restricts mating choices
- Possibility that an offspring will inherit two copies of the same ancestral allele
- Define F, the inbreeding coefficient, as the probability that a random individual in the population inherits two copies of the same allele from a common ancestor
Admixture as a Confounder
- Consider the problem of estimating the effect of a SNP on a disease phenotype: \beta in P(Y=1) = \text{logit}^{-1}(\alpha + \beta G)
- Recent admixture mixes ancestries within individuals: genotype is a convex combination of source populations
- If phenotype prevalence differs by ancestry, local or global ancestry proportions act like hidden covariates
- Association tests must separate causal signal from ancestry-driven allele frequency differences
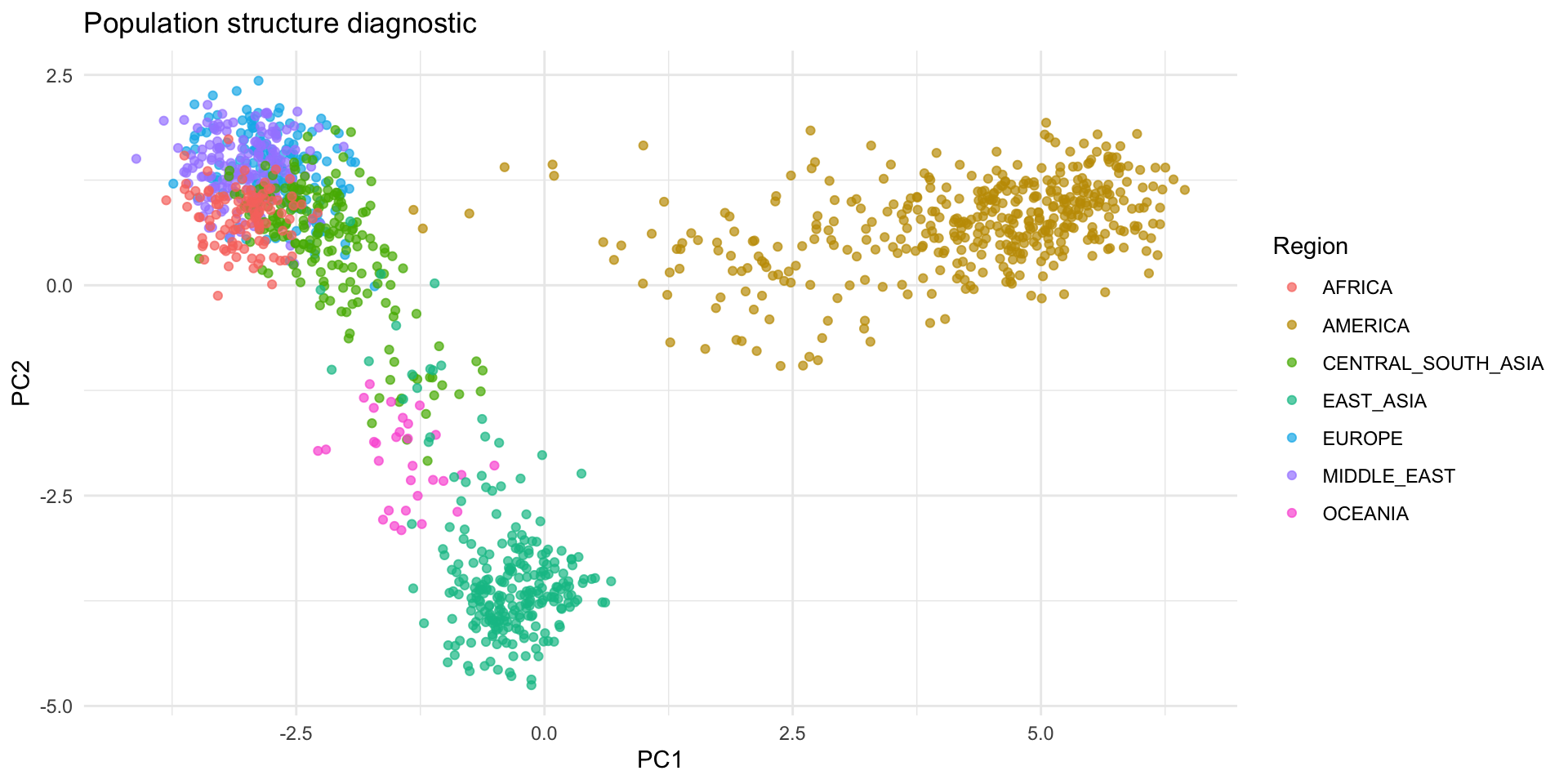
Principal Components for Structure
- Construct the standardized genotype matrix Z and compute Z^T Z / M (with M markers)
- Top eigenvectors capture major ancestry gradients
- Use the leading PCs as covariates in association tests or to stratify downstream analyses
Principal Components for Structure
/// GENIND OBJECT /////////
// 1,350 individuals; 678 loci; 8,170 alleles; size: 44.1 Mb
// Basic content
@tab: 1350 x 8170 matrix of allele counts
@loc.n.all: number of alleles per locus (range: 5-35)
@loc.fac: locus factor for the 8170 columns of @tab
@all.names: list of allele names for each locus
@ploidy: ploidy of each individual (range: 2-2)
@type: codom
@call: read.fstat(file = file, missing = missing, quiet = quiet)
// Optional content
@pop: population of each individual (group size range: 3-50)
@other: a list containing: popInfo Principal Components for Structure
hgdp_df <- genind2df(eHGDP, sep = "/")
# Convert to allele count matrix, center columns (replace missing with locus means)
geno_mat <- scaleGen(eHGDP, center = TRUE, scale = FALSE, NA.method = "mean")
pc_fit <- prcomp(geno_mat, center = FALSE, scale. = FALSE)
# Map individuals to geographic regions for coloring
pop_info <- eHGDP@other$popInfo
pop_index <- as.integer(pop(eHGDP))
region <- pop_info$Region[pop_index]
plot_df <- data.frame(PC1 = pc_fit$x[, 1], PC2 = pc_fit$x[, 2], Region = region)
ggplot(plot_df, aes(PC1, PC2, color = Region)) +
geom_point(alpha = 0.7, size = 1.5) +
labs(title = "Population structure diagnostic", x = "PC1", y = "PC2") +
theme_minimal()Principal Components for Structure
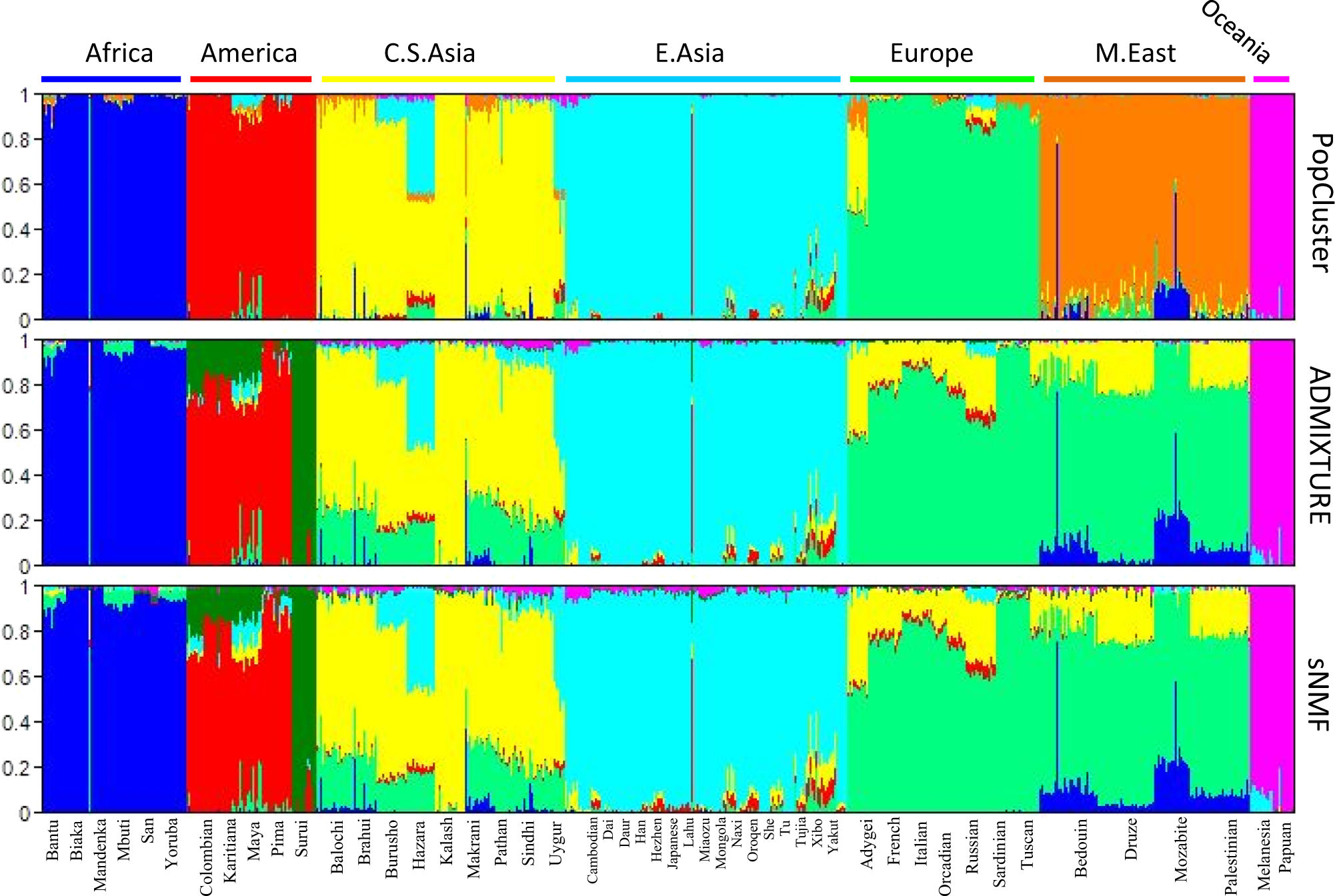
STRUCTURE & ADMIXTURE
- Model-based clustering methods for population structure inference
- Assume K latent populations with distinct allele frequencies
- Each individual has ancestry proportions \boldsymbol{\pi}_i across K populations
- Each genotype is drawn from a mixture of population-specific allele frequencies
- Use maximum likelihood (ADMIXTURE) or Bayesian inference (STRUCTURE) to estimate parameters
Assumptions
- Hardy-Weinberg equilibrium within each ancestral population
- Linkage equilibrium between loci within each ancestral population
- Loci are unlinked (or weakly linked)
Example Output
Bayesian Admixture
- Latent populations k = 1,\ldots,K possess allele frequencies \theta_{k\ell} at locus \ell
- Individual ancestry proportions \boldsymbol{\pi}_i \sim \text{Dirichlet}(\boldsymbol{\alpha})
- Genotype y_{i\ell} \sim \text{Binomial}\left(2, \sum_{k} \pi_{ik} \theta_{k\ell}\right) assuming HWE within each ancestral population
- Posterior draws propagate ancestry/allele-frequency uncertainty into association testing, local ancestry, and polygenic prediction
Identifiability & Label Switching
- Problem: Mixture components are exchangeable → posterior multimodality
- Solution: Anchor loci with informative priors break symmetry
- Populations defined by genetic signatures, not arbitrary labels
- Multiple anchors provide robustness against weak signals
Stan Model: Data & Parameters
data {
int<lower=1> N; // individuals
int<lower=1> L; // loci
int<lower=1> K; // ancestral pops
array[N, L] int<lower=0, upper=2> y; // genotypes (0,1,2)
int<lower=0> L_soft;
array[L_soft] int<lower=1, upper=L> soft_idx; // e.g., {2}
array[K] real<lower=0> a_theta; // Beta 'a' for non-anchor loci
array[K] real<lower=0> b_theta; // Beta 'b' for non-anchor loci
int<lower=1, upper=L> l_star; // anchor locus index
real<lower=0> conc_pi; // shared Dirichlet conc. for pi
}Stan Model: Parameters & Transformed Parameters
parameters {
array[N] simplex[K] pi; // ancestry proportions
ordered[K] eta; // ordered logits at anchor locus
matrix<lower=0, upper=1>[K, L-1] theta_rest; // allele freqs for non-anchor loci
}
transformed parameters {
matrix<lower=0, upper=1>[K, L] theta; // full allele-frequency matrix
matrix[N, L] p_mix; // mixed allele frequency
// anchor column (ordered)
for (k in 1:K) theta[k, l_star] = inv_logit(eta[k]);
// fill remaining columns
{
int c = 1;
for (l in 1:L) {
if (l == l_star) continue;
for (k in 1:K) theta[k, l] = theta_rest[k, c];
c += 1;
}
}
// mixture expectations
for (n in 1:N)
for (l in 1:L)
p_mix[n, l] = dot_product(pi[n], col(theta, l));
}Stan Model: Priors & Likelihood
model {
// priors
eta ~ normal(0, 2.5); // weak prior; ordering gives ID
for (k in 1:K)
for (c in 1:(L - 1))
theta_rest[k, c] ~ beta(a_theta[k], b_theta[k]);
for (n in 1:N)
pi[n] ~ dirichlet(rep_vector(conc_pi, K));
for (c in 1:L_soft) {
int l = soft_idx[c];
if (l != l_star) {
// comp 1 LOW, comp 2 HIGH at these loci (gentle)
target += beta_lpdf(theta[1, l] | 2, 8);
target += beta_lpdf(theta[2, l] | 8, 2);
}
}
// likelihood
for (n in 1:N)
for (l in 1:L)
y[n, l] ~ binomial(2, p_mix[n, l]);
}Stan Model: Generated Quantities
Mathematical Framework
Individual-Locus Allele Frequency
For individual i at locus \ell, expected allele frequency: p_{i\ell} = \sum_{k=1}^K \pi_{ik} \theta_{k\ell}
Information Borrowing Across Loci
Key insight: Same \boldsymbol{\pi}_i parameters appear in likelihood for all loci
L(\boldsymbol{\pi}_i, \boldsymbol{\Theta}) = \prod_{\ell=1}^L \text{Binomial}\left(y_{i\ell} \mid 2, \sum_{k=1}^K \pi_{ik} \theta_{k\ell}\right)
Mathematical Framework
Hierarchical Learning
- Anchor loci provide strong identification signal
- Remaining loci contribute cumulative evidence
- Posterior uncertainty propagates through all parameters
Simulating Admixed Genotypes
- 1
- Fixed random seed for consistent results across sessions
- 2
- Normalized gamma variates generate simplex-constrained ancestry proportions
- 3
- Set simulation parameters
Simulating Admixed Genotypes
4# Create very clear population differentiation
theta_true <- matrix(0, nrow = K, ncol = L)
# Multiple anchor loci: very strong differentiation
theta_true[1, 1] <- 0.1 # Pop 1: very low frequency at anchor 1
theta_true[2, 1] <- 0.9 # Pop 2: very high frequency at anchor 1
theta_true[1, 2] <- 0.1 # Pop 1: very low frequency at anchor 2
theta_true[2, 2] <- 0.9 # Pop 2: very high frequency at anchor 2
# Other loci: strong differentiation
theta_true[1, 3:L] <- rbeta(L-2, 2, 8) # Pop 1: much lower overall
theta_true[2, 3:L] <- rbeta(L-2, 8, 2) # Pop 2: much higher overall
5pi_true <- rbind(
rdirichlet(12, c(90, 10)), # pop 1
rdirichlet(12, c(10, 90)), # Very pure pop 2
rdirichlet(16, c(10, 10)) # Admixed individuals
)- 4
- Strong genetic signatures with multiple anchor loci
- 5
- Mix of unadmixed founders and admixed descendants
Simulating Admixed Genotypes
6y <- matrix(0L, nrow = N, ncol = L)
for (n in 1:N) {
for (l in 1:L) {
p_mix <- sum(pi_true[n, ] * theta_true[, l])
y[n, l] <- rbinom(1, size = 2, prob = p_mix)
}
}- 6
- Genotypes drawn from Binomial(2, p_mix) per individual and locus
PCA On Simulated Genotypes
- Center genotypes by 2 \times \hat{p} and scale by \sqrt{2 \cdot \hat{p} \cdot (1 - \hat{p})}
p_hat <- colMeans(y) / 2
sd_hat <- sqrt(pmax(1e-6, 2 * p_hat * (1 - p_hat)))
Z <- scale(y, center = 2 * p_hat, scale = sd_hat)
pc <- prcomp(Z, center = FALSE, scale. = FALSE)
pc_df <- data.frame(PC1 = pc$x[, 1], PC2 = pc$x[, 2], pi1 = pi_true[, 1])
ggplot(pc_df, aes(PC1, PC2, color = pi1)) +
geom_point(alpha = 0.7, size = 1.6) +
scale_color_viridis_c(end = .8) +
labs(color = expression(pi[1]), x = "PC1", y = "PC2") +
geom_vline(xintercept = 2.75, linetype = "dashed", color = "gray40", linewidth = 0.6) +
geom_vline(xintercept = -2.75, linetype = "dashed", color = "gray40", linewidth = 0.6) +
annotate("text", x = 4.5, y = 6, label = "Pop 1", color = "gray20", size = 6) +
annotate("text", x = -4.5, y = 6, label = "Pop 2", color = "gray20", size = 6) +
annotate("text", x = 0, y = 6, label = "Admixed", color = "gray20", size = 6) +
theme_bw(base_size = 14) +
theme(panel.grid = element_blank()) PCA On Simulated Genotypes
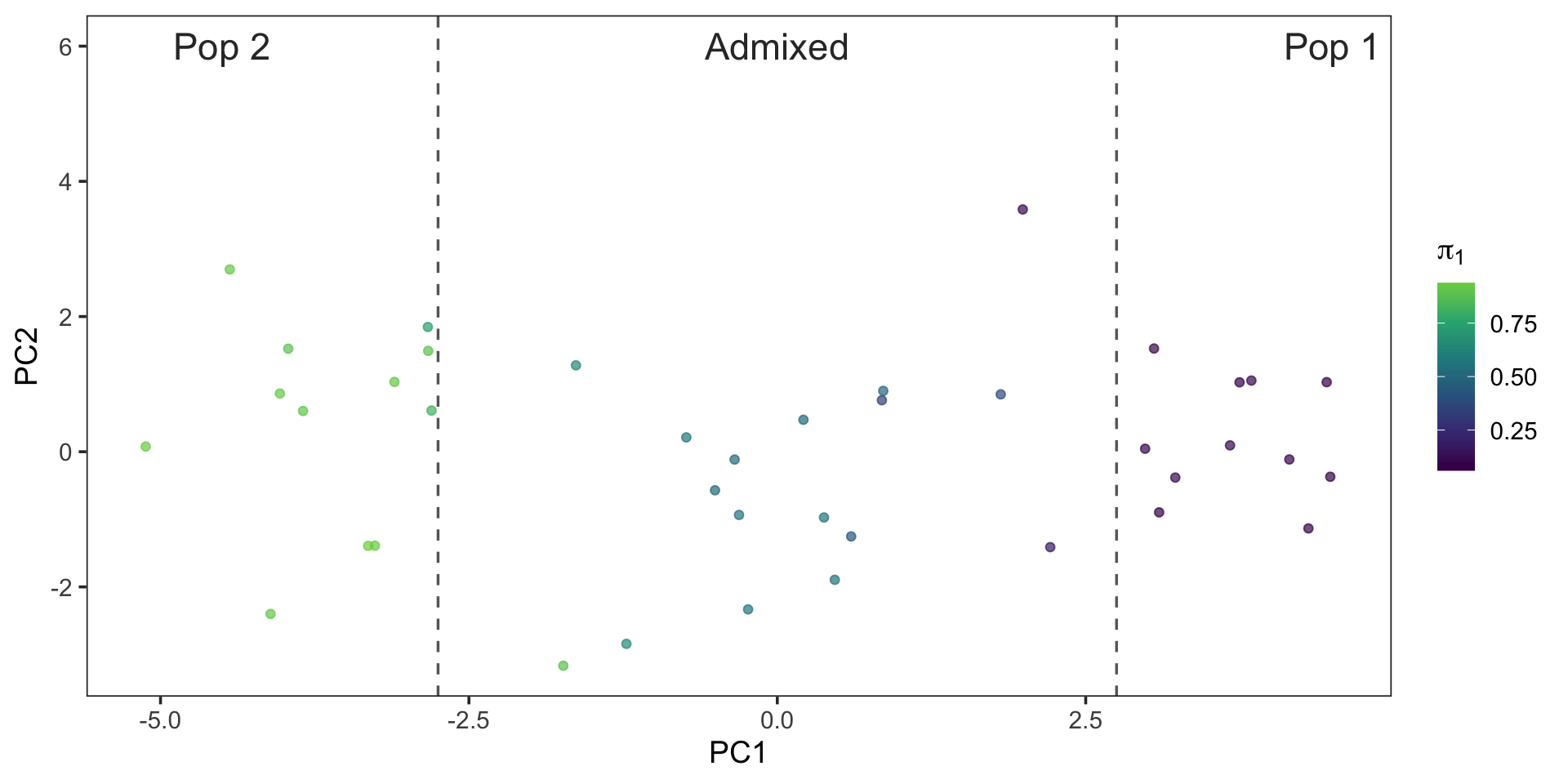
Fitting the Model
admix_mod <- cmdstan_model(file.path("stan", "structure_admixture.stan"))
l_star <- 1 # choose your anchor locus (e.g., 1)
admix_data <- list(
N = N, L = L, K = K, y = y,
a_theta = rep(1.5, K), b_theta = rep(1.5, K),
l_star = l_star, L_soft = 1, soft_idx = c(2),
conc_pi = 2.0
)
admix_fit <- admix_mod$sample(
data = admix_data,
chains = 4, parallel_chains = 4,
iter_warmup = 2000, iter_sampling = 2000,
seed = 887804, refresh = 0
)Running MCMC with 4 parallel chains...Chain 2 Informational Message: The current Metropolis proposal is about to be rejected because of the following issue:Chain 2 Exception: binomial_lpmf: Probability parameter is 1, but must be in the interval [0, 1] (in '/var/folders/3f/7lk7ddbn19j4f1rzxtlx3z9w0000gn/T/RtmpX9Wapd/model-d8207aefdca0.stan', line 65, column 6 to column 41)Chain 2 If this warning occurs sporadically, such as for highly constrained variable types like covariance matrices, then the sampler is fine,Chain 2 but if this warning occurs often then your model may be either severely ill-conditioned or misspecified.Chain 2 Chain 4 Informational Message: The current Metropolis proposal is about to be rejected because of the following issue:Chain 4 Exception: binomial_lpmf: Probability parameter is 1, but must be in the interval [0, 1] (in '/var/folders/3f/7lk7ddbn19j4f1rzxtlx3z9w0000gn/T/RtmpX9Wapd/model-d8207aefdca0.stan', line 65, column 6 to column 41)Chain 4 If this warning occurs sporadically, such as for highly constrained variable types like covariance matrices, then the sampler is fine,Chain 4 but if this warning occurs often then your model may be either severely ill-conditioned or misspecified.Chain 4 Chain 2 finished in 8.6 seconds.
Chain 1 finished in 8.7 seconds.
Chain 4 finished in 8.6 seconds.
Chain 3 finished in 9.0 seconds.
All 4 chains finished successfully.
Mean chain execution time: 8.7 seconds.
Total execution time: 9.1 seconds.Stan Output
dm <- admix_fit$draws(variables = c("theta","pi"), format = "draws_df")
lstar <- admix_data$l_star
L <- ncol(y)
non_anchor <- setdiff(1:L, lstar)
# Orientation score per draw: average sign over non-anchor loci
sign_per_draw <- rowMeans(sapply(non_anchor, function(l)
sign(dm[[sprintf("theta[2,%d]", l)]] - dm[[sprintf("theta[1,%d]", l)]])))
# Reference orientation: the majority sign across all draws
ref_sign <- ifelse(mean(sign_per_draw) >= 0, -1, 1)
# Decide which DRAWS to flip (not just which chains)
flip_draw <- sign_per_draw * ref_sign < 0
# Helper: swap theta rows and pi columns for those draws
swap_block <- function(df, pat1, pat2, idx){
i1 <- grep(pat1, names(df)); i2 <- grep(pat2, names(df))
tmp <- df[idx, i1, drop=FALSE]
df[idx, i1] <- df[idx, i2, drop=FALSE]
df[idx, i2] <- tmp
df
}
# Swap theta rows
dm <- swap_block(dm, "^theta\\[1,", "^theta\\[2,", flip_draw)
# Swap pi columns
dm <- swap_block(dm, "^pi\\[[0-9]+,1\\]$", "^pi\\[[0-9]+,2\\]$", flip_draw)Aligned Posteriors
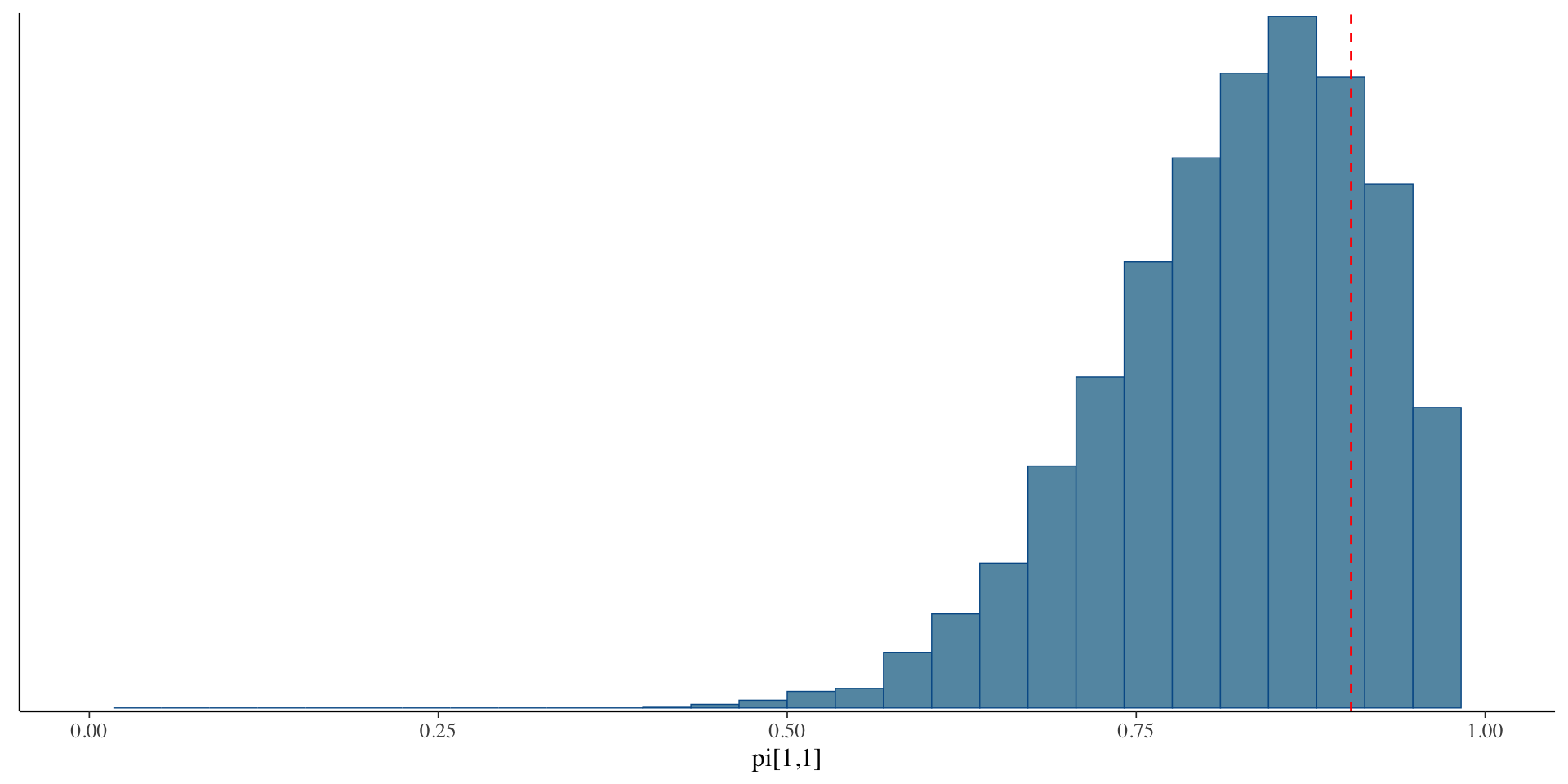
Credible Intervals
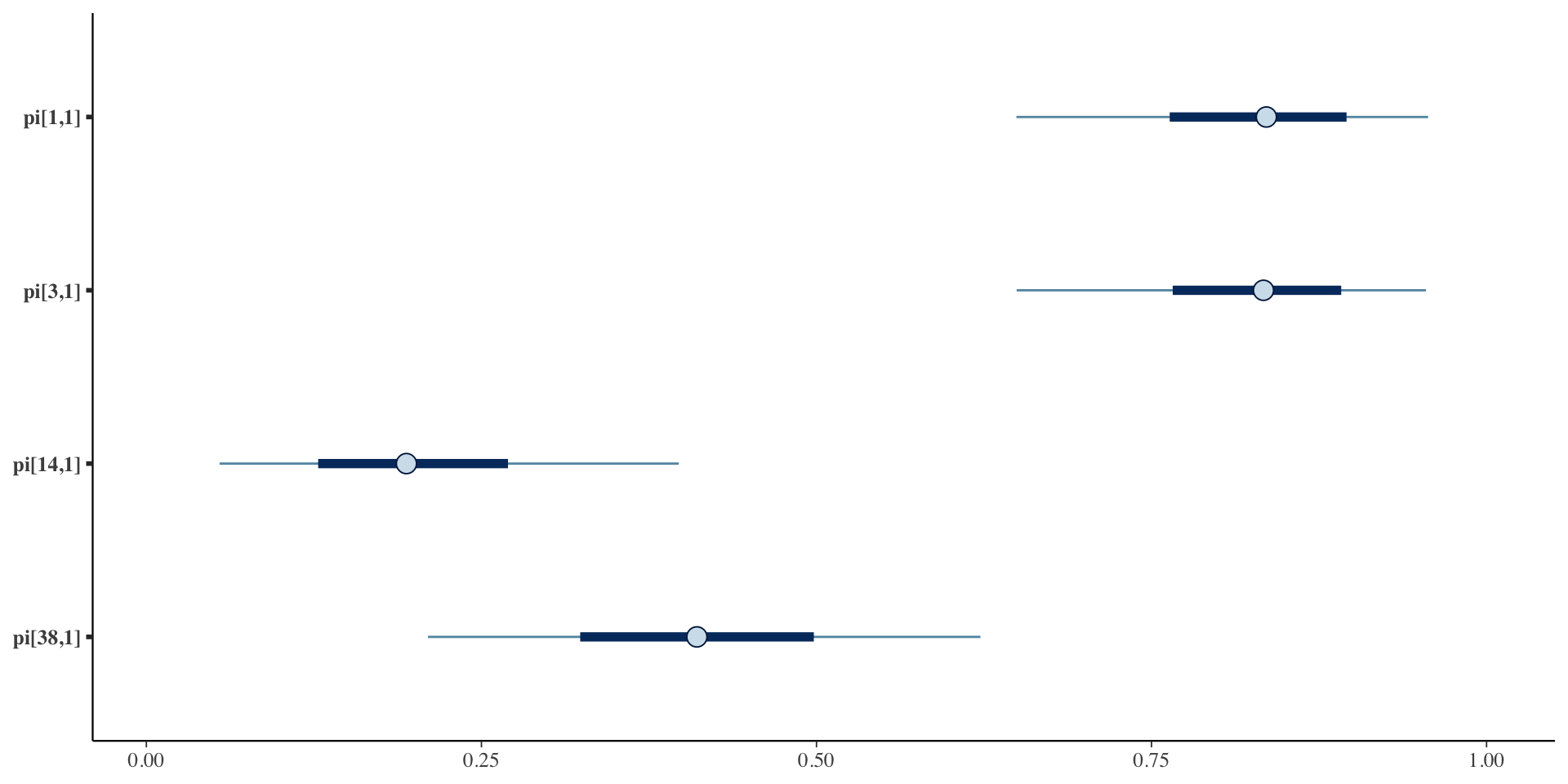
Summary & Next Steps
- Population structure influences genetic analyses
- Bayesian framing exposes prior choices, enables posterior uncertainty on structure (ABO example, admixture Stan model)
- Bayesian modeling as attempting to capture the data-generating process