HAPHPIPE Installation Instructions
HAPHIPE depends on more than a dozen different programs, each of which may itself depend on other programs and libraries. Installing everything separately would be a nightmare, so you should use the package manager "Bioconda" to install the HAPHPIPE package. Bioconda is a popular package manager in the bioinformatics world. See the Helpful Resources section for more information and resources for Bioconda. Here, we will describe where to get Bioconda and how to install it, then how to use Bioconda to install HAPHPIPE. We will also detail the acquisition and installation of one program, GATK, that is not handled by Bioconda.
Here, we describe the procedure for installing HAPHPIPE using the package manager Bioconda (Grüning et al. 2018) on the command line. If you are unfamiliar with Bioconda, please see for installation and channel setup. The source code for HAPHPIPE is available here and is written in Python 3.7.2.
The installation process begins with the creation of a conda environment that installs HAPHPIPE and its dependencies together with one short command. Once the conda environment has been created, it can be activated for use with the command conda activate haphpipe. Due to license restrictions, Bioconda cannot distribute and install GATK (McKenna et al. 2010; Van der Auwera et al. 2013; Poplin et al. 2018) directly. To fully install GATK, you must download a licensed copy of GATK (version 3.8-0) from the Broad Institute (link).
A video showing the full HAPHPIPE installation process and the automatic demo is available here.
1. Install conda
Download the conda package:
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-MacOSX-x86_64.sh
sh Miniconda3-latest-MacOSX-x86_64.sh
2. Set up conda channels
These need to be put in as a command in the order they come, as this sets the priority for where packages are pulled from. Therefore, in this order, conda-forge is top priority and explored first when downloading a program.
conda config --add channels R
conda config --add channels defaults
conda config --add channels bioconda
conda config --add channels conda-forge
3. Create a conda environment for HAPHPIPE
conda create -n haphpipe haphpipe
This will install HAPHPIPE and all dependencies in Bioconda.Note: on some HPC systems (including GW's Pegasus), certain dependencies must be installed as a 'User Install'. If this is the case (which will be apparent if you receive availability errors for any of the above packages after installing them through conda) use the following command to install the package:
pip install --user <package name>
4. Activate the environment
conda activate haphpipe
You can now test your HAPHPIPE install by running haphpipe -h.
5. Install GATK
Due to license restrictions, bioconda cannot distribute
and install GATK directly. To fully install GATK, you must
download a licensed copy of GATK (version 3.8-0) from the Broad Institute and register the package using gatk3-register. gsutil has been included as a dependency in HAPHPIPE, so you do not need to worry about installing it for the GATK acquisition.
This will copy GATK into your conda environment.
GATK needs to be installed directly on the command line with the following commands:
mkdir -p /path/to/gatk_dir
gsutil cp gs://gatk-software/package-archive/gatk/GenomeAnalysisTK-3.8-0-ge9d806836.tar.bz2 path/to/gatk_dir/
gatk3-register /path/to/gatk_dir/GenomeAnalysisTK.jar
NOTE: HAPHPIPE was developed and tested using GATK 3.8.
PredictHaplo Installation Instructions
Users are required to download PredictHaplo on their own computing system prior to running any of the haplotype modules (predict_haplo and ph_parser).
Here is how the GW CBI team installed PredictHaplo onto our HPC, which has a slurm scheduling system and uses Lua module files.
We cannot help with the installation of this software, but have provided the code that we used here to install PredictHaplo onto our system. Please see their website for contact information if needed.
This module loads predicthaplo onto GWU's HPC - Colonial One.
See https://bmda.dmi.unibas.ch/software.html
cd /path/to/modules/predicthaplo
# use gcc 4.9.4, add blas and lapack to library path
module load gcc/4.9.4
module load blas/gcc/64
module load lapack/gcc/64
# download source
cd archive
wget https://bmda.dmi.unibas.ch/software/PredictHaplo-Paired-0.4.tgz
cd ..
# unzip source and change directory name
tar xfvz archive/PredictHaplo-Paired-0.4.tgz
mv PredictHaplo-Paired-0.4 0.4
cd 0.4
# install scythestat
tar xfvz scythestat-1.0.3.tar.gz
cd scythestat-1.0.3
./configure --prefix=/path/to/modules/predicthaplo/0.4/NEWSCYTHE
make install
cd ..
# compile predicthaplo
make
If a segfault error occurs during the hp_predict_haplo module, this is not a characteristic of HAPHPIPE but rather that of PredictHaplo. Sometimes, we have luck if we just rerun the code again or move to an interactive CPU node. We are unsure what causes this error, and we only see it between the local and global reconstruction phases in PredictHaplo.
Quick-Start
1. Activate haphpipe
Make sure you have conda running.
For students at GW using Colonial One, you need to load the miniconda3 module like such prior to activating the haphpipe conda environemnt:
module load miniconda3.
conda activate haphpipe
2. Test that it is loaded correctly
haphpipe -h
should produce:
Program: haphpipe (haplotype and phylodynamics pipeline)
Version: 1.0.2
Commands:
-- Reads
sample_reads subsample reads using seqtk
trim_reads trim reads using Trimmomatic
join_reads join reads using FLASh
ec_reads error correct reads using SPAdes
-- Assemble
assemble_denovo assemble reads denovo
assemble_amplicons assemble contigs to amplicon regions
assemble_scaffold assemble contigs to genome
align_reads align reads to reference
call_variants call variants
vcf_to_consensus create consensus sequence from VCF
refine_assembly iterative refinement: align - variants - consensus
finalize_assembly finalize consensus sequence
-- Haplotype
predict_haplo assemble haplotypes with PredictHaplo
ph_parser parse output from PredictHaplo
cliquesnv assemble haplotypes with CliqueSNV
-- Description
pairwise_align align consensus to an annotated reference
extract_pairwise extract sequence regions from pairwise alignment
summary_stats generates summary statistics for samples
-- Phylo
multiple_align multiple sequence alignment
model_test tests for model of evolution using ModelTest
build_tree_NG builds phylogenetic tree with RAxML-NG
-- Miscellaneous
demo setup demo directory and test data
Directory Structure
Below is the recommended directory structure for using HAPHPIPE:
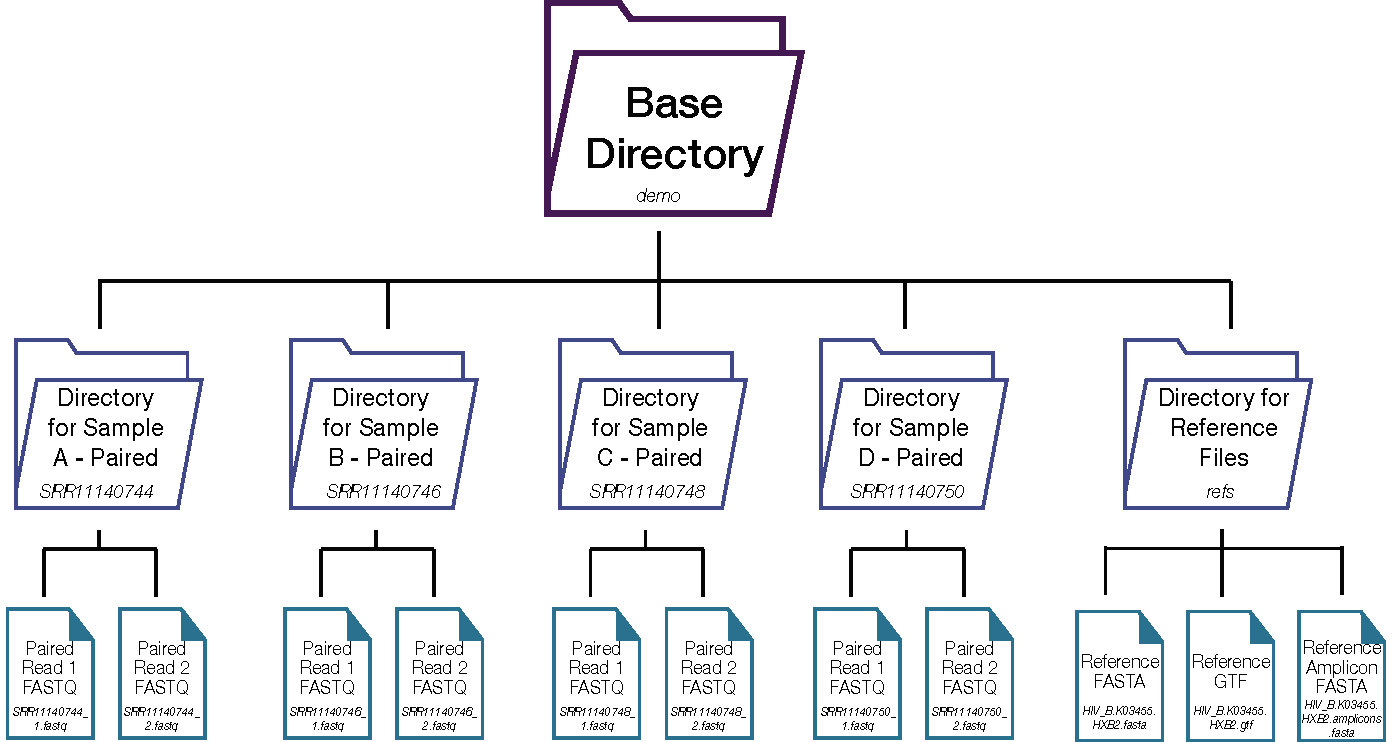
We recommend creating a separate directory for each sample, as well as a directory for reference files.
Windows Users
HAPHPIPE is only available for Mac OSX or Linux platforms. We suggest the following options for running HAPHPIPE on a Windows machine:
-
Run HAPHPIPE on your institution's HPC cluster, if available.
-
Utilize the Windows Subsystem for Linux.
-
Run Linux in a virtual machine via VirtualBox
Reference Files
Several modules in HAPHPIPE use reference files: either a FASTA file containing a reference sequence or a GTF file denoting genome regions for amplicon assembly. With HIV data, we use
the file HIV_B.K03455.HXB2.fasta as a reference whole-genome file, HIV_B.K03455.HXB2.amplicons.fasta for amplicon assembly, and HIV_B.K03455.HXB2.gtf as a GTF file. All three files are downloaded in the demo module.
The contents of each file are below.
HIV_B.K03455.HXB2.fasta
>HIV_B.K03455.HXB2
TGGAAGGGCTAATTCACTCCCAACGAAGACAAGATATCCTTGATCTGTGGATCTACCACACACAAGGCTACTTCCCTGATTAGCAGAACTACACACCAGG
GCCAGGGATCAGATATCCACTGACCTTTGGATGGTGCTACAAGCTAGTACCAGTTGAGCCAGAGAAGTTAGAAGAAGCCAACAAAGGAGAGAACACCAGC
TTGTTACACCCTGTGAGCCTGCATGGAATGGATGACCCGGAGAGAGAAGTGTTAGAGTGGAGGTTTGACAGCCGCCTAGCATTTCATCACATGGCCCGAG
AGCTGCATCCGGAGTACTTCAAGAACTGCTGACATCGAGCTTGCTACAAGGGACTTTCCGCTGGGGACTTTCCAGGGAGGCGTGGCCTGGGCGGGACTGG
GGAGTGGCGAGCCCTCAGATCCTGCATATAAGCAGCTGCTTTTTGCCTGTACTGGGTCTCTCTGGTTAGACCAGATCTGAGCCTGGGAGCTCTCTGGCTA
ACTAGGGAACCCACTGCTTAAGCCTCAATAAAGCTTGCCTTGAGTGCTTCAAGTAGTGTGTGCCCGTCTGTTGTGTGACTCTGGTAACTAGAGATCCCTC
AGACCCTTTTAGTCAGTGTGGAAAATCTCTAGCAGTGGCGCCCGAACAGGGACCTGAAAGCGAAAGGGAAACCAGAGGAGCTCTCTCGACGCAGGACTCG
GCTTGCTGAAGCGCGCACGGCAAGAGGCGAGGGGCGGCGACTGGTGAGTACGCCAAAAATTTTGACTAGCGGAGGCTAGAAGGAGAGAGATGGGTGCGAG
AGCGTCAGTATTAAGCGGGGGAGAATTAGATCGATGGGAAAAAATTCGGTTAAGGCCAGGGGGAAAGAAAAAATATAAATTAAAACATATAGTATGGGCA
AGCAGGGAGCTAGAACGATTCGCAGTTAATCCTGGCCTGTTAGAAACATCAGAAGGCTGTAGACAAATACTGGGACAGCTACAACCATCCCTTCAGACAG
GATCAGAAGAACTTAGATCATTATATAATACAGTAGCAACCCTCTATTGTGTGCATCAAAGGATAGAGATAAAAGACACCAAGGAAGCTTTAGACAAGAT
AGAGGAAGAGCAAAACAAAAGTAAGAAAAAAGCACAGCAAGCAGCAGCTGACACAGGACACAGCAATCAGGTCAGCCAAAATTACCCTATAGTGCAGAAC
ATCCAGGGGCAAATGGTACATCAGGCCATATCACCTAGAACTTTAAATGCATGGGTAAAAGTAGTAGAAGAGAAGGCTTTCAGCCCAGAAGTGATACCCA
TGTTTTCAGCATTATCAGAAGGAGCCACCCCACAAGATTTAAACACCATGCTAAACACAGTGGGGGGACATCAAGCAGCCATGCAAATGTTAAAAGAGAC
CATCAATGAGGAAGCTGCAGAATGGGATAGAGTGCATCCAGTGCATGCAGGGCCTATTGCACCAGGCCAGATGAGAGAACCAAGGGGAAGTGACATAGCA
GGAACTACTAGTACCCTTCAGGAACAAATAGGATGGATGACAAATAATCCACCTATCCCAGTAGGAGAAATTTATAAAAGATGGATAATCCTGGGATTAA
ATAAAATAGTAAGAATGTATAGCCCTACCAGCATTCTGGACATAAGACAAGGACCAAAGGAACCCTTTAGAGACTATGTAGACCGGTTCTATAAAACTCT
AAGAGCCGAGCAAGCTTCACAGGAGGTAAAAAATTGGATGACAGAAACCTTGTTGGTCCAAAATGCGAACCCAGATTGTAAGACTATTTTAAAAGCATTG
GGACCAGCGGCTACACTAGAAGAAATGATGACAGCATGTCAGGGAGTAGGAGGACCCGGCCATAAGGCAAGAGTTTTGGCTGAAGCAATGAGCCAAGTAA
CAAATTCAGCTACCATAATGATGCAGAGAGGCAATTTTAGGAACCAAAGAAAGATTGTTAAGTGTTTCAATTGTGGCAAAGAAGGGCACACAGCCAGAAA
TTGCAGGGCCCCTAGGAAAAAGGGCTGTTGGAAATGTGGAAAGGAAGGACACCAAATGAAAGATTGTACTGAGAGACAGGCTAATTTTTTAGGGAAGATC
TGGCCTTCCTACAAGGGAAGGCCAGGGAATTTTCTTCAGAGCAGACCAGAGCCAACAGCCCCACCAGAAGAGAGCTTCAGGTCTGGGGTAGAGACAACAA
CTCCCCCTCAGAAGCAGGAGCCGATAGACAAGGAACTGTATCCTTTAACTTCCCTCAGGTCACTCTTTGGCAACGACCCCTCGTCACAATAAAGATAGGG
GGGCAACTAAAGGAAGCTCTATTAGATACAGGAGCAGATGATACAGTATTAGAAGAAATGAGTTTGCCAGGAAGATGGAAACCAAAAATGATAGGGGGAA
TTGGAGGTTTTATCAAAGTAAGACAGTATGATCAGATACTCATAGAAATCTGTGGACATAAAGCTATAGGTACAGTATTAGTAGGACCTACACCTGTCAA
CATAATTGGAAGAAATCTGTTGACTCAGATTGGTTGCACTTTAAATTTTCCCATTAGCCCTATTGAGACTGTACCAGTAAAATTAAAGCCAGGAATGGAT
GGCCCAAAAGTTAAACAATGGCCATTGACAGAAGAAAAAATAAAAGCATTAGTAGAAATTTGTACAGAGATGGAAAAGGAAGGGAAAATTTCAAAAATTG
GGCCTGAAAATCCATACAATACTCCAGTATTTGCCATAAAGAAAAAAGACAGTACTAAATGGAGAAAATTAGTAGATTTCAGAGAACTTAATAAGAGAAC
TCAAGACTTCTGGGAAGTTCAATTAGGAATACCACATCCCGCAGGGTTAAAAAAGAAAAAATCAGTAACAGTACTGGATGTGGGTGATGCATATTTTTCA
GTTCCCTTAGATGAAGACTTCAGGAAGTATACTGCATTTACCATACCTAGTATAAACAATGAGACACCAGGGATTAGATATCAGTACAATGTGCTTCCAC
AGGGATGGAAAGGATCACCAGCAATATTCCAAAGTAGCATGACAAAAATCTTAGAGCCTTTTAGAAAACAAAATCCAGACATAGTTATCTATCAATACAT
GGATGATTTGTATGTAGGATCTGACTTAGAAATAGGGCAGCATAGAACAAAAATAGAGGAGCTGAGACAACATCTGTTGAGGTGGGGACTTACCACACCA
GACAAAAAACATCAGAAAGAACCTCCATTCCTTTGGATGGGTTATGAACTCCATCCTGATAAATGGACAGTACAGCCTATAGTGCTGCCAGAAAAAGACA
GCTGGACTGTCAATGACATACAGAAGTTAGTGGGGAAATTGAATTGGGCAAGTCAGATTTACCCAGGGATTAAAGTAAGGCAATTATGTAAACTCCTTAG
AGGAACCAAAGCACTAACAGAAGTAATACCACTAACAGAAGAAGCAGAGCTAGAACTGGCAGAAAACAGAGAGATTCTAAAAGAACCAGTACATGGAGTG
TATTATGACCCATCAAAAGACTTAATAGCAGAAATACAGAAGCAGGGGCAAGGCCAATGGACATATCAAATTTATCAAGAGCCATTTAAAAATCTGAAAA
CAGGAAAATATGCAAGAATGAGGGGTGCCCACACTAATGATGTAAAACAATTAACAGAGGCAGTGCAAAAAATAACCACAGAAAGCATAGTAATATGGGG
AAAGACTCCTAAATTTAAACTGCCCATACAAAAGGAAACATGGGAAACATGGTGGACAGAGTATTGGCAAGCCACCTGGATTCCTGAGTGGGAGTTTGTT
AATACCCCTCCCTTAGTGAAATTATGGTACCAGTTAGAGAAAGAACCCATAGTAGGAGCAGAAACCTTCTATGTAGATGGGGCAGCTAACAGGGAGACTA
AATTAGGAAAAGCAGGATATGTTACTAATAGAGGAAGACAAAAAGTTGTCACCCTAACTGACACAACAAATCAGAAGACTGAGTTACAAGCAATTTATCT
AGCTTTGCAGGATTCGGGATTAGAAGTAAACATAGTAACAGACTCACAATATGCATTAGGAATCATTCAAGCACAACCAGATCAAAGTGAATCAGAGTTA
GTCAATCAAATAATAGAGCAGTTAATAAAAAAGGAAAAGGTCTATCTGGCATGGGTACCAGCACACAAAGGAATTGGAGGAAATGAACAAGTAGATAAAT
TAGTCAGTGCTGGAATCAGGAAAGTACTATTTTTAGATGGAATAGATAAGGCCCAAGATGAACATGAGAAATATCACAGTAATTGGAGAGCAATGGCTAG
TGATTTTAACCTGCCACCTGTAGTAGCAAAAGAAATAGTAGCCAGCTGTGATAAATGTCAGCTAAAAGGAGAAGCCATGCATGGACAAGTAGACTGTAGT
CCAGGAATATGGCAACTAGATTGTACACATTTAGAAGGAAAAGTTATCCTGGTAGCAGTTCATGTAGCCAGTGGATATATAGAAGCAGAAGTTATTCCAG
CAGAAACAGGGCAGGAAACAGCATATTTTCTTTTAAAATTAGCAGGAAGATGGCCAGTAAAAACAATACATACTGACAATGGCAGCAATTTCACCGGTGC
TACGGTTAGGGCCGCCTGTTGGTGGGCGGGAATCAAGCAGGAATTTGGAATTCCCTACAATCCCCAAAGTCAAGGAGTAGTAGAATCTATGAATAAAGAA
TTAAAGAAAATTATAGGACAGGTAAGAGATCAGGCTGAACATCTTAAGACAGCAGTACAAATGGCAGTATTCATCCACAATTTTAAAAGAAAAGGGGGGA
TTGGGGGGTACAGTGCAGGGGAAAGAATAGTAGACATAATAGCAACAGACATACAAACTAAAGAATTACAAAAACAAATTACAAAAATTCAAAATTTTCG
GGTTTATTACAGGGACAGCAGAAATCCACTTTGGAAAGGACCAGCAAAGCTCCTCTGGAAAGGTGAAGGGGCAGTAGTAATACAAGATAATAGTGACATA
AAAGTAGTGCCAAGAAGAAAAGCAAAGATCATTAGGGATTATGGAAAACAGATGGCAGGTGATGATTGTGTGGCAAGTAGACAGGATGAGGATTAGAACA
TGGAAAAGTTTAGTAAAACACCATATGTATGTTTCAGGGAAAGCTAGGGGATGGTTTTATAGACATCACTATGAAAGCCCTCATCCAAGAATAAGTTCAG
AAGTACACATCCCACTAGGGGATGCTAGATTGGTAATAACAACATATTGGGGTCTGCATACAGGAGAAAGAGACTGGCATTTGGGTCAGGGAGTCTCCAT
AGAATGGAGGAAAAAGAGATATAGCACACAAGTAGACCCTGAACTAGCAGACCAACTAATTCATCTGTATTACTTTGACTGTTTTTCAGACTCTGCTATA
AGAAAGGCCTTATTAGGACACATAGTTAGCCCTAGGTGTGAATATCAAGCAGGACATAACAAGGTAGGATCTCTACAATACTTGGCACTAGCAGCATTAA
TAACACCAAAAAAGATAAAGCCACCTTTGCCTAGTGTTACGAAACTGACAGAGGATAGATGGAACAAGCCCCAGAAGACCAAGGGCCACAGAGGGAGCCA
CACAATGAATGGACACTAGAGCTTTTAGAGGAGCTTAAGAATGAAGCTGTTAGACATTTTCCTAGGATTTGGCTCCATGGCTTAGGGCAACATATCTATG
AAACTTATGGGGATACTTGGGCAGGAGTGGAAGCCATAATAAGAATTCTGCAACAACTGCTGTTTATCCATTTTCAGAATTGGGTGTCGACATAGCAGAA
TAGGCGTTACTCGACAGAGGAGAGCAAGAAATGGAGCCAGTAGATCCTAGACTAGAGCCCTGGAAGCATCCAGGAAGTCAGCCTAAAACTGCTTGTACCA
ATTGCTATTGTAAAAAGTGTTGCTTTCATTGCCAAGTTTGTTTCATAACAAAAGCCTTAGGCATCTCCTATGGCAGGAAGAAGCGGAGACAGCGACGAAG
AGCTCATCAGAACAGTCAGACTCATCAAGCTTCTCTATCAAAGCAGTAAGTAGTACATGTAACGCAACCTATACCAATAGTAGCAATAGTAGCATTAGTA
GTAGCAATAATAATAGCAATAGTTGTGTGGTCCATAGTAATCATAGAATATAGGAAAATATTAAGACAAAGAAAAATAGACAGGTTAATTGATAGACTAA
TAGAAAGAGCAGAAGACAGTGGCAATGAGAGTGAAGGAGAAATATCAGCACTTGTGGAGATGGGGGTGGAGATGGGGCACCATGCTCCTTGGGATGTTGA
TGATCTGTAGTGCTACAGAAAAATTGTGGGTCACAGTCTATTATGGGGTACCTGTGTGGAAGGAAGCAACCACCACTCTATTTTGTGCATCAGATGCTAA
AGCATATGATACAGAGGTACATAATGTTTGGGCCACACATGCCTGTGTACCCACAGACCCCAACCCACAAGAAGTAGTATTGGTAAATGTGACAGAAAAT
TTTAACATGTGGAAAAATGACATGGTAGAACAGATGCATGAGGATATAATCAGTTTATGGGATCAAAGCCTAAAGCCATGTGTAAAATTAACCCCACTCT
GTGTTAGTTTAAAGTGCACTGATTTGAAGAATGATACTAATACCAATAGTAGTAGCGGGAGAATGATAATGGAGAAAGGAGAGATAAAAAACTGCTCTTT
CAATATCAGCACAAGCATAAGAGGTAAGGTGCAGAAAGAATATGCATTTTTTTATAAACTTGATATAATACCAATAGATAATGATACTACCAGCTATAAG
TTGACAAGTTGTAACACCTCAGTCATTACACAGGCCTGTCCAAAGGTATCCTTTGAGCCAATTCCCATACATTATTGTGCCCCGGCTGGTTTTGCGATTC
TAAAATGTAATAATAAGACGTTCAATGGAACAGGACCATGTACAAATGTCAGCACAGTACAATGTACACATGGAATTAGGCCAGTAGTATCAACTCAACT
GCTGTTAAATGGCAGTCTAGCAGAAGAAGAGGTAGTAATTAGATCTGTCAATTTCACGGACAATGCTAAAACCATAATAGTACAGCTGAACACATCTGTA
GAAATTAATTGTACAAGACCCAACAACAATACAAGAAAAAGAATCCGTATCCAGAGAGGACCAGGGAGAGCATTTGTTACAATAGGAAAAATAGGAAATA
TGAGACAAGCACATTGTAACATTAGTAGAGCAAAATGGAATAACACTTTAAAACAGATAGCTAGCAAATTAAGAGAACAATTTGGAAATAATAAAACAAT
AATCTTTAAGCAATCCTCAGGAGGGGACCCAGAAATTGTAACGCACAGTTTTAATTGTGGAGGGGAATTTTTCTACTGTAATTCAACACAACTGTTTAAT
AGTACTTGGTTTAATAGTACTTGGAGTACTGAAGGGTCAAATAACACTGAAGGAAGTGACACAATCACCCTCCCATGCAGAATAAAACAAATTATAAACA
TGTGGCAGAAAGTAGGAAAAGCAATGTATGCCCCTCCCATCAGTGGACAAATTAGATGTTCATCAAATATTACAGGGCTGCTATTAACAAGAGATGGTGG
TAATAGCAACAATGAGTCCGAGATCTTCAGACCTGGAGGAGGAGATATGAGGGACAATTGGAGAAGTGAATTATATAAATATAAAGTAGTAAAAATTGAA
CCATTAGGAGTAGCACCCACCAAGGCAAAGAGAAGAGTGGTGCAGAGAGAAAAAAGAGCAGTGGGAATAGGAGCTTTGTTCCTTGGGTTCTTGGGAGCAG
CAGGAAGCACTATGGGCGCAGCCTCAATGACGCTGACGGTACAGGCCAGACAATTATTGTCTGGTATAGTGCAGCAGCAGAACAATTTGCTGAGGGCTAT
TGAGGCGCAACAGCATCTGTTGCAACTCACAGTCTGGGGCATCAAGCAGCTCCAGGCAAGAATCCTGGCTGTGGAAAGATACCTAAAGGATCAACAGCTC
CTGGGGATTTGGGGTTGCTCTGGAAAACTCATTTGCACCACTGCTGTGCCTTGGAATGCTAGTTGGAGTAATAAATCTCTGGAACAGATTTGGAATCACA
CGACCTGGATGGAGTGGGACAGAGAAATTAACAATTACACAAGCTTAATACACTCCTTAATTGAAGAATCGCAAAACCAGCAAGAAAAGAATGAACAAGA
ATTATTGGAATTAGATAAATGGGCAAGTTTGTGGAATTGGTTTAACATAACAAATTGGCTGTGGTATATAAAATTATTCATAATGATAGTAGGAGGCTTG
GTAGGTTTAAGAATAGTTTTTGCTGTACTTTCTATAGTGAATAGAGTTAGGCAGGGATATTCACCATTATCGTTTCAGACCCACCTCCCAACCCCGAGGG
GACCCGACAGGCCCGAAGGAATAGAAGAAGAAGGTGGAGAGAGAGACAGAGACAGATCCATTCGATTAGTGAACGGATCCTTGGCACTTATCTGGGACGA
TCTGCGGAGCCTGTGCCTCTTCAGCTACCACCGCTTGAGAGACTTACTCTTGATTGTAACGAGGATTGTGGAACTTCTGGGACGCAGGGGGTGGGAAGCC
CTCAAATATTGGTGGAATCTCCTACAGTATTGGAGTCAGGAACTAAAGAATAGTGCTGTTAGCTTGCTCAATGCCACAGCCATAGCAGTAGCTGAGGGGA
CAGATAGGGTTATAGAAGTAGTACAAGGAGCTTGTAGAGCTATTCGCCACATACCTAGAAGAATAAGACAGGGCTTGGAAAGGATTTTGCTATAAGATGG
GTGGCAAGTGGTCAAAAAGTAGTGTGATTGGATGGCCTACTGTAAGGGAAAGAATGAGACGAGCTGAGCCAGCAGCAGATAGGGTGGGAGCAGCATCTCG
AGACCTGGAAAAACATGGAGCAATCACAAGTAGCAATACAGCAGCTACCAATGCTGCTTGTGCCTGGCTAGAAGCACAAGAGGAGGAGGAGGTGGGTTTT
CCAGTCACACCTCAGGTACCTTTAAGACCAATGACTTACAAGGCAGCTGTAGATCTTAGCCACTTTTTAAAAGAAAAGGGGGGACTGGAAGGGCTAATTC
ACTCCCAAAGAAGACAAGATATCCTTGATCTGTGGATCTACCACACACAAGGCTACTTCCCTGATTAGCAGAACTACACACCAGGGCCAGGGGTCAGATA
TCCACTGACCTTTGGATGGTGCTACAAGCTAGTACCAGTTGAGCCAGATAAGATAGAAGAGGCCAATAAAGGAGAGAACACCAGCTTGTTACACCCTGTG
AGCCTGCATGGGATGGATGACCCGGAGAGAGAAGTGTTAGAGTGGAGGTTTGACAGCCGCCTAGCATTTCATCACGTGGCCCGAGAGCTGCATCCGGAGT
ACTTCAAGAACTGCTGACATCGAGCTTGCTACAAGGGACTTTCCGCTGGGGACTTTCCAGGGAGGCGTGGCCTGGGCGGGACTGGGGAGTGGCGAGCCCT
CAGATCCTGCATATAAGCAGCTGCTTTTTGCCTGTACTGGGTCTCTCTGGTTAGACCAGATCTGAGCCTGGGAGCTCTCTGGCTAACTAGGGAACCCACT
GCTTAAGCCTCAATAAAGCTTGCCTTGAGTGCTTCAAGTAGTGTGTGCCCGTCTGTTGTGTGACTCTGGTAACTAGAGATCCCTCAGACCCTTTTAGTCA
GTGTGGAAAATCTCTAGCA
HIV_B.K03455.HXB2.amplicons.fasta
>ref|HIV_B.K03455.HXB2|reg|PRRT|
CCCTCAGGTCACTCTTTGGCAACGACCCCTCGTCACAATAAAGATAGGGGGGCAACTAAAGGAAGCTCTATTAGATACAG
GAGCAGATGATACAGTATTAGAAGAAATGAGTTTGCCAGGAAGATGGAAACCAAAAATGATAGGGGGAATTGGAGGTTTT
ATCAAAGTAAGACAGTATGATCAGATACTCATAGAAATCTGTGGACATAAAGCTATAGGTACAGTATTAGTAGGACCTAC
ACCTGTCAACATAATTGGAAGAAATCTGTTGACTCAGATTGGTTGCACTTTAAATTTTCCCATTAGCCCTATTGAGACTG
TACCAGTAAAATTAAAGCCAGGAATGGATGGCCCAAAAGTTAAACAATGGCCATTGACAGAAGAAAAAATAAAAGCATTA
GTAGAAATTTGTACAGAGATGGAAAAGGAAGGGAAAATTTCAAAAATTGGGCCTGAAAATCCATACAATACTCCAGTATT
TGCCATAAAGAAAAAAGACAGTACTAAATGGAGAAAATTAGTAGATTTCAGAGAACTTAATAAGAGAACTCAAGACTTCT
GGGAAGTTCAATTAGGAATACCACATCCCGCAGGGTTAAAAAAGAAAAAATCAGTAACAGTACTGGATGTGGGTGATGCA
TATTTTTCAGTTCCCTTAGATGAAGACTTCAGGAAGTATACTGCATTTACCATACCTAGTATAAACAATGAGACACCAGG
GATTAGATATCAGTACAATGTGCTTCCACAGGGATGGAAAGGATCACCAGCAATATTCCAAAGTAGCATGACAAAAATCT
TAGAGCCTTTTAGAAAACAAAATCCAGACATAGTTATCTATCAATACATGGATGATTTGTATGTAGGATCTGACTTAGAA
ATAGGGCAGCATAGAACAAAAATAGAGGAGCTGAGACAACATCTGTTGAGGTGGGGACTTACCACACCAGACAAAAAACA
TCAGAAAGAACCTCCATTCCTTTGGATGGGTTATGAACTCCATCCTGATAAATGGACAGTACAGCCTATAGTGCTGCCAG
AAAAAGACAGCTGGACTGTCAATGACATACAGAAGTTAGTGGGGAAATTGAATTGGGCAAGTCAGATTTACCCAGGGATT
AAAGTAAGGCAATTATGTAAACTCCTTAGAGGAACCAAAGCACTAACAGAAGTAATACCACTAACAGAAGAAGCAGAGCT
AGAACTGGCAGAAAACAGAGAGATTCTAAAAGAACCAGTACATGGAGTGTATTATGACCCATCAAAAGACTTAATAGCAG
AAATACAGAAGCAGGGGCAAGGCCAATGGACATATCAAATTTATCAAGAGCCATTTAAAAATCTGAAAACAGGAAAATAT
GCAAGAATGAGGGGTGCCCACACTAATGATGTAAAACAATTAACAGAGGCAGTGCAAAAAATAACCACAGAAAGCATAGT
AATATGGGGAAAGACTCCTAAATTTAAACTGCCCATACAAAAGGAAACATGGGAAACATGGTGGACAGAGTATTGGCAAG
CCACCTGGATTCCTGAGTGGGAGTTTGTTAATACCCCTCCCTTAGTGAAATTATGGTACCAGTTAGAGAAAGAACCCATA
GTAGGAGCAGAAACCTTC
>ref|HIV_B.K03455.HXB2|reg|INT|
TTTTTAGATGGAATAGATAAGGCCCAAGATGAACATGAGAAATATCACAGTAATTGGAGAGCAATGGCTAGTGATTTTAA
CCTGCCACCTGTAGTAGCAAAAGAAATAGTAGCCAGCTGTGATAAATGTCAGCTAAAAGGAGAAGCCATGCATGGACAAG
TAGACTGTAGTCCAGGAATATGGCAACTAGATTGTACACATTTAGAAGGAAAAGTTATCCTGGTAGCAGTTCATGTAGCC
AGTGGATATATAGAAGCAGAAGTTATTCCAGCAGAAACAGGGCAGGAAACAGCATATTTTCTTTTAAAATTAGCAGGAAG
ATGGCCAGTAAAAACAATACATACTGACAATGGCAGCAATTTCACCGGTGCTACGGTTAGGGCCGCCTGTTGGTGGGCGG
GAATCAAGCAGGAATTTGGAATTCCCTACAATCCCCAAAGTCAAGGAGTAGTAGAATCTATGAATAAAGAATTAAAGAAA
ATTATAGGACAGGTAAGAGATCAGGCTGAACATCTTAAGACAGCAGTACAAATGGCAGTATTCATCCACAATTTTAAAAG
AAAAGGGGGGATTGGGGGGTACAGTGCAGGGGAAAGAATAGTAGACATAATAGCAACAGACATACAAACTAAAGAATTAC
AAAAACAAATTACAAAAATTCAAAATTTTCGGGTTTATTACAGGGACAGCAGAAATCCACTTTGGAAAGGACCAGCAAAG
CTCCTCTGGAAAGGTGAAGGGGCAGTAGTAATACAAGATAATAGTGACATAAAAGTAGTGCCAAGAAGAAAAGCAAAGAT
CATTAGGGATTATGGAAAACAGATGGCAGGTGATGATTGTGTGGCAAGTAGACAGGATGAGGAT
>ref|HIV_B.K03455.HXB2|reg|gp120|
CAGTAGATCCTAGACTAGAGCCCTGGAAGCATCCAGGAAGTCAGCCTAAAACTGCTTGTACCAATTGCTATTGTAAAAAG
TGTTGCTTTCATTGCCAAGTTTGTTTCATAACAAAAGCCTTAGGCATCTCCTATGGCAGGAAGAAGCGGAGACAGCGACG
AAGAGCTCATCAGAACAGTCAGACTCATCAAGCTTCTCTATCAAAGCAGTAAGTAGTACATGTAACGCAACCTATACCAA
TAGTAGCAATAGTAGCATTAGTAGTAGCAATAATAATAGCAATAGTTGTGTGGTCCATAGTAATCATAGAATATAGGAAA
ATATTAAGACAAAGAAAAATAGACAGGTTAATTGATAGACTAATAGAAAGAGCAGAAGACAGTGGCAATGAGAGTGAAGG
AGAAATATCAGCACTTGTGGAGATGGGGGTGGAGATGGGGCACCATGCTCCTTGGGATGTTGATGATCTGTAGTGCTACA
GAAAAATTGTGGGTCACAGTCTATTATGGGGTACCTGTGTGGAAGGAAGCAACCACCACTCTATTTTGTGCATCAGATGC
TAAAGCATATGATACAGAGGTACATAATGTTTGGGCCACACATGCCTGTGTACCCACAGACCCCAACCCACAAGAAGTAG
TATTGGTAAATGTGACAGAAAATTTTAACATGTGGAAAAATGACATGGTAGAACAGATGCATGAGGATATAATCAGTTTA
TGGGATCAAAGCCTAAAGCCATGTGTAAAATTAACCCCACTCTGTGTTAGTTTAAAGTGCACTGATTTGAAGAATGATAC
TAATACCAATAGTAGTAGCGGGAGAATGATAATGGAGAAAGGAGAGATAAAAAACTGCTCTTTCAATATCAGCACAAGCA
TAAGAGGTAAGGTGCAGAAAGAATATGCATTTTTTTATAAACTTGATATAATACCAATAGATAATGATACTACCAGCTAT
AAGTTGACAAGTTGTAACACCTCAGTCATTACACAGGCCTGTCCAAAGGTATCCTTTGAGCCAATTCCCATACATTATTG
TGCCCCGGCTGGTTTTGCGATTCTAAAATGTAATAATAAGACGTTCAATGGAACAGGACCATGTACAAATGTCAGCACAG
TACAATGTACACATGGAATTAGGCCAGTAGTATCAACTCAACTGCTGTTAAATGGCAGTCTAGCAGAAGAAGAGGTAGTA
ATTAGATCTGTCAATTTCACGGACAATGCTAAAACCATAATAGTACAGCTGAACACATCTGTAGAAATTAATTGTACAAG
ACCCAACAACAATACAAGAAAAAGAATCCGTATCCAGAGAGGACCAGGGAGAGCATTTGTTACAATAGGAAAAATAGGAA
ATATGAGACAAGCACATTGTAACATTAGTAGAGCAAAATGGAATAACACTTTAAAACAGATAGCTAGCAAATTAAGAGAA
CAATTTGGAAATAATAAAACAATAATCTTTAAGCAATCCTCAGGAGGGGACCCAGAAATTGTAACGCACAGTTTTAATTG
TGGAGGGGAATTTTTCTACTGTAATTCAACACAACTGTTTAATAGTACTTGGTTTAATAGTACTTGGAGTACTGAAGGGT
CAAATAACACTGAAGGAAGTGACACAATCACCCTCCCATGCAGAATAAAACAAATTATAAACATGTGGCAGAAAGTAGGA
AAAGCAATGTATGCCCCTCCCATCAGTGGACAAATTAGATGTTCATCAAATATTACAGGGCTGCTATTAACAAGAGATGG
TGGTAATAGCAACAATGAGTCCGAGATCTTCAGACCTGGAGGAGGAGATATGAGGGACAATTGGAGAAGTGAATTATATA
AATATAAAGTAGTAAAAATTGAACCATTAGGAGTAGCACCCACCAAGGCAAAGAGAAGAGTGGTGCAGAGAGAAAAAAGA
HIV_B.K03455.HXB2.gtf
HIV_B.K03455.HXB2 LANL amplicon 2252 3869 . + 2 name "PRRT"; primary_cds "2252-2549"; alt_cds "2550-3869";
HIV_B.K03455.HXB2 LANL amplicon 4230 5093 . + 0 name "INT"; primary_cds "2085-5096"; alt_cds "5098-5619";
HIV_B.K03455.HXB2 LANL amplicon 6225 7757 . + 1 name "gp120"; primary_cds "6225-8795"; alt_cds "5831-6223";
